5GXF
 
 | |
5GXE
 
 | |
4GY5
 
 | | Crystal structure of the tandem tudor domain and plant homeodomain of UHRF1 with Histone H3K9me3 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Peptide from Histone H3.3, ZINC ION | | Authors: | Cheng, J, Yang, Y, Fang, J, Xiao, J, Zhu, T, Chen, F, Wang, P, Xu, Y. | | Deposit date: | 2012-09-05 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.956 Å) | | Cite: | Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein
J.Biol.Chem., 288, 2013
|
|
7XR8
 
 | |
5Z2S
 
 | | Crystal structure of DUX4-HD2 domain | | Descriptor: | Double homeobox protein 4 | | Authors: | Dong, X, Zhang, W, Wu, H, Huang, J, Zhang, M, Wang, P, Zhang, H, Chen, Z, Chen, S, Meng, G. | | Deposit date: | 2018-01-03 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of DUX4/IGH-driven transactivation.
Leukemia, 32, 2018
|
|
5Z2T
 
 | | Crystal structure of DNA-bound DUX4-HD2 | | Descriptor: | 5'-D(*TP*TP*CP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*TP*T)-3', 5'-D(P*AP*AP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*GP*T)-3', Double homeobox protein 4 | | Authors: | Dong, X, Zhang, W, Wu, H, Huang, J, Zhang, M, Wang, P, Zhang, H, Chen, Z, Chen, S, Meng, G. | | Deposit date: | 2018-01-04 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.623 Å) | | Cite: | Structural basis of DUX4/IGH-driven transactivation.
Leukemia, 32, 2018
|
|
6AK1
 
 | | Crystal structure of DmoA from Hyphomicrobium sulfonivorans | | Descriptor: | Dimethyl-sulfide monooxygenase | | Authors: | Cao, H.Y, Wang, P, Peng, M, Li, C.Y. | | Deposit date: | 2018-08-28 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.284 Å) | | Cite: | Crystal structure of the dimethylsulfide monooxygenase DmoA from Hyphomicrobium sulfonivorans.
Acta Crystallogr.,Sect.F, 74, 2018
|
|
5XZD
 
 | |
6IMQ
 
 | | Crystal structure of PML B1-box multimers | | Descriptor: | CHLORIDE ION, Protein PML, ZINC ION | | Authors: | Li, Y, Ma, X, Chen, Z, Wu, H, Wang, P, Wu, W, Cheng, N, Zeng, L, Zhang, H, Cai, X, Chen, S.J, Chen, Z, Meng, G. | | Deposit date: | 2018-10-23 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | B1 oligomerization regulates PML nuclear body biogenesis and leukemogenesis.
Nat Commun, 10, 2019
|
|
6IJC
 
 | | Structure of MMPA-CoA dehydrogenase from Roseovarius nubinhibens ISM | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acyl-CoA dehydrogenase family protein | | Authors: | Shao, X, Yuan, Z.L, Cao, H.Y, Wang, P, Li, C.Y, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2018-10-09 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insight into 3-methylmercaptopropionate metabolism and kinetical regulation of demethylation pathway in marine dimethylsulfoniopropionate-catabolizing bacteria.
Mol.Microbiol., 111, 2019
|
|
7KPL
 
 | | Crystal structure of hEphB1 in apo form | | Descriptor: | Ephrin type-B receptor 1 | | Authors: | Ahmed, M, Wang, P, Sadek, H. | | Deposit date: | 2020-11-11 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Identification of tetracycline combinations as EphB1 tyrosine kinase inhibitors for treatment of neuropathic pain.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KPM
 
 | | Crystal structure of hEphB1 bound with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ephrin type-B receptor 1 | | Authors: | Ahmed, M, Wang, P, Sadek, H. | | Deposit date: | 2020-11-11 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.608 Å) | | Cite: | Identification of tetracycline combinations as EphB1 tyrosine kinase inhibitors for treatment of neuropathic pain.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6IHK
 
 | | Structure of MMPA CoA ligase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AMP-binding domain protein | | Authors: | Shao, X, Cao, H.Y, Wang, P, Li, C.Y, Zhao, F, Peng, M, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2018-09-30 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Mechanistic insight into 3-methylmercaptopropionate metabolism and kinetical regulation of demethylation pathway in marine dimethylsulfoniopropionate-catabolizing bacteria.
Mol.Microbiol., 111, 2019
|
|
6IJB
 
 | | Structure of 3-methylmercaptopropionate CoA ligase mutant K523A in complex with AMP and MMPA | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(methylsulfanyl)propanoic acid, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Shao, X, Cao, H.Y, Wang, P, Li, C.Y, Zhao, F, Peng, M, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2018-10-09 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.111 Å) | | Cite: | Mechanistic insight into 3-methylmercaptopropionate metabolism and kinetical regulation of demethylation pathway in marine dimethylsulfoniopropionate-catabolizing bacteria.
Mol.Microbiol., 111, 2019
|
|
7E2W
 
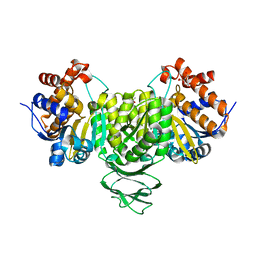 | | Crystal structure of isocitrate dehydrogenase from Ostreococcus tauri in complex with isocitrate and magnesium(II) | | Descriptor: | CITRATE ANION, GLYCEROL, ISOCITRIC ACID, ... | | Authors: | Zhu, G.P, Tang, W.G, Wang, P. | | Deposit date: | 2021-02-07 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of NAD + -linked isocitrate dehydrogenase from the green alga Ostreococcus tauri and its evolutionary relationship with eukaryotic NADP + -linked homologs.
Arch.Biochem.Biophys., 708, 2021
|
|
7C8G
 
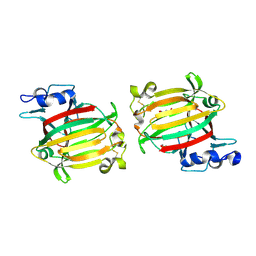 | | Structure of alginate lyase AlyC3 | | Descriptor: | Alginate lyase AlyC3, GLYCEROL, SUCCINIC ACID | | Authors: | Zhang, Y.Z, Xu, F, Chen, X.L, Wang, P. | | Deposit date: | 2020-05-30 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and molecular basis for the substrate positioning mechanism of a new PL7 subfamily alginate lyase from the arctic.
J.Biol.Chem., 295, 2020
|
|
7C8F
 
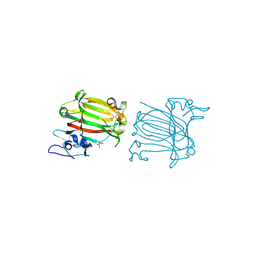 | | Structure of alginate lyase AlyC3 in complex with dimannuronate(2M) | | Descriptor: | H127A/Y244A mutant of alginate lyase AlyC3 in complex with dimannuronate, MALONATE ION, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Zhang, Y.Z, Xu, F, Chen, X.L, Wang, P. | | Deposit date: | 2020-05-30 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.461 Å) | | Cite: | Structural and molecular basis for the substrate positioning mechanism of a new PL7 subfamily alginate lyase from the arctic.
J.Biol.Chem., 295, 2020
|
|
7M8K
 
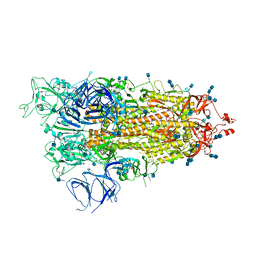 | | Cryo-EM structure of Brazil (P.1) SARS-CoV-2 spike glycoprotein variant in the prefusion state (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Casner, R.G, Cerutti, G, Shapiro, L, Ho, D.D. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-05 | | Last modified: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY | | Cite: | Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization.
Cell Host Microbe, 29, 2021
|
|
7N5H
 
 | | Cryo-EM structure of broadly neutralizing antibody 2-36 in complex with prefusion SARS-CoV-2 spike glycoprotein | | Descriptor: | 2-36 Fab heavy chain, 2-36 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Casner, R.G, Cerutti, G, Shapiro, L. | | Deposit date: | 2021-06-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | A monoclonal antibody that neutralizes SARS-CoV-2 variants, SARS-CoV, and other sarbecoviruses.
Emerg Microbes Infect, 11, 2022
|
|
7S4G
 
 | | Fab fragment bound to the Cter peptide of Ly6G6D | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Lymphocyte antigen 6 complex locus protein G6d, ... | | Authors: | Rouge, L, Lupardus, P. | | Deposit date: | 2021-09-08 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel Anti-LY6G6D/CD3 T-Cell-Dependent Bispecific Antibody for the Treatment of Colorectal Cancer.
Mol.Cancer Ther., 21, 2022
|
|
7VP9
 
 | | Crystal structure of human ClpP in complex with ZG111 | | Descriptor: | (6S,9aS)-N-[(4-bromophenyl)methyl]-6-[(2S)-butan-2-yl]-8-(naphthalen-1-ylmethyl)-4,7-bis(oxidanylidene)-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, mitochondrial, ... | | Authors: | Wang, P.Y, Gan, J.H, Yang, C.-G. | | Deposit date: | 2021-10-15 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Aberrant human ClpP activation disturbs mitochondrial proteome homeostasis to suppress pancreatic ductal adenocarcinoma.
Cell Chem Biol, 29, 2022
|
|
1FHR
 
 | | SOLUTION STRUCTURE OF THE FHA2 DOMAIN OF RAD53 COMPLEXED WITH A PHOSPHOTYROSYL PEPTIDE | | Descriptor: | DNA REPAIR PROTEIN RAD9, PROTEIN KINASE SPK1 | | Authors: | Byeon, I.-J.L, Liao, H, Yongkiettrakul, S, Tsai, M.-D. | | Deposit date: | 2000-08-02 | | Release date: | 2000-10-18 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | II. Structure and specificity of the interaction between the FHA2 domain of Rad53 and phosphotyrosyl peptides.
J.Mol.Biol., 302, 2000
|
|
1FHQ
 
 | |
4PY4
 
 | |
7RX6
 
 | |
