6KD9
 
 | | Binding pose 1 of 2-CF3 bound AsqJ complex | | Descriptor: | (3~{Z})-4-methyl-3-[[4-(trifluoromethyl)phenyl]methylidene]-1~{H}-1,4-benzodiazepine-2,5-dione, FE (III) ION, Iron/alpha-ketoglutarate-dependent dioxygenase asqJ, ... | | Authors: | Liao, H.J, Chan, N.L. | | Deposit date: | 2019-07-01 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Binding pose 1 of 2-CF3 bound AsqJ complex
J.Am.Chem.Soc., 2020
|
|
5Y7R
 
 | | Quaternary complex of AsqJ-Fe3+-2OG-cyclopeptin | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Iron/alpha-ketoglutarate-dependent dioxygenase asqJ, ... | | Authors: | Liao, H.J, Lin, T.S. | | Deposit date: | 2017-08-17 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Insights into the Desaturation of Cyclopeptin and its C3 Epimer Catalyzed by a non-Heme Iron Enzyme: Structural Characterization and Mechanism Elucidation.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6K0E
 
 | | The quinary complex of AsqJ-Fe-2OG-dehydrocyclopeptin-dioxygen | | Descriptor: | (3~{Z})-4-methyl-3-(phenylmethylidene)-1~{H}-1,4-benzodiazepine-2,5-dione, 2-OXOGLUTARIC ACID, FE (III) ION, ... | | Authors: | Liao, H.J, Chan, N.L. | | Deposit date: | 2019-05-06 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | The quinary complex of AsqJ-Fe-2OG-dehydrocyclopeptin-dioxygen
J.Am.Chem.Soc., 2020
|
|
6K0F
 
 | | The crystal structure of cyclopenin-bound AsqJ quinary complex | | Descriptor: | (3~{R},3'~{S})-4-methyl-3'-phenyl-spiro[1~{H}-1,4-benzodiazepine-3,2'-oxirane]-2,5-dione, 2-OXOGLUTARIC ACID, FE (III) ION, ... | | Authors: | Liao, H.J, Chan, N.L. | | Deposit date: | 2019-05-06 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.634 Å) | | Cite: | The crystal structure of cyclopenin-bound AsqJ quinary complex
J.Am.Chem.Soc., 2020
|
|
5Y7T
 
 | | Quaternary complex of AsqJ-Fe3+-2OG-D-cyclopeptin | | Descriptor: | (3R)-4-methyl-3-(phenylmethyl)-1,3-dihydro-1,4-benzodiazepine-2,5-dione, 2-OXOGLUTARIC ACID, FE (III) ION, ... | | Authors: | Liao, H.J. | | Deposit date: | 2017-08-18 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Insights into the Desaturation of Cyclopeptin and its C3 Epimer Catalyzed by a non-Heme Iron Enzyme: Structural Characterization and Mechanism Elucidation.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6JZM
 
 | | The intermediate forming O-C10 bond formation in AsqJ-catalyzed epoxidation | | Descriptor: | (3~{R},3'~{S})-4-methyl-3'-phenyl-spiro[1~{H}-1,4-benzodiazepine-3,2'-oxirane]-2,5-dione, FE (III) ION, Iron/alpha-ketoglutarate-dependent dioxygenase asqJ, ... | | Authors: | Liao, H.J, Chan, N.L. | | Deposit date: | 2019-05-02 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The intermediate forming O-C10 bond formation in AsqJ-catalyzed epoxidation
J.Am.Chem.Soc., 2020
|
|
6K30
 
 | | Binding pose 2 of 2-CF3 bound AsqJ complex | | Descriptor: | (3~{Z})-4-methyl-3-[[4-(trifluoromethyl)phenyl]methylidene]-1~{H}-1,4-benzodiazepine-2,5-dione, FE (III) ION, Iron/alpha-ketoglutarate-dependent dioxygenase asqJ, ... | | Authors: | Liao, H.J, Chan, N.L. | | Deposit date: | 2019-05-15 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.684 Å) | | Cite: | Binding pose 2 of 2-CF3 bound AsqJ complex
J.Am.Chem.Soc., 2020
|
|
6KV2
 
 | |
7XAZ
 
 | | Crystal structure of Omicron BA.1.1 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liao, H, Meng, Y, Li, W. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
1QU5
 
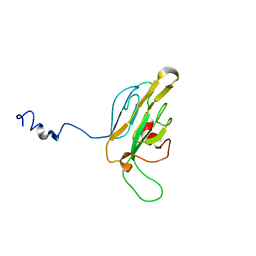 | |
4V6Q
 
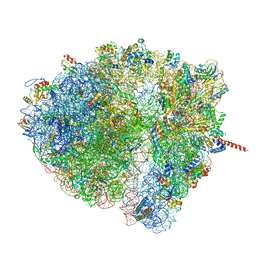 | | Structural characterization of mRNA-tRNA translocation intermediates (class 5 of the six classes) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Liao, H, Schreiner, E, Fu, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-12-08 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Structural characterization of mRNA-tRNA translocation intermediates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4V6R
 
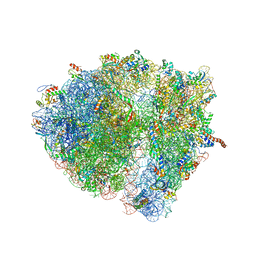 | | Structural characterization of mRNA-tRNA translocation intermediates (class 6 of the six classes) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Liao, H, Schreiner, E, Fu, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-12-08 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Structural characterization of mRNA-tRNA translocation intermediates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4V6N
 
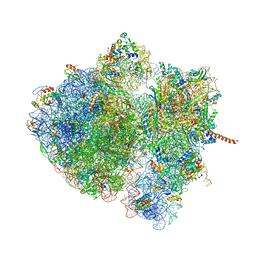 | | Structural characterization of mRNA-tRNA translocation intermediates (50S ribosome of class2 of the six classes) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Liao, H, Schreiner, E, Fu, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (12.1 Å) | | Cite: | Structural characterization of mRNA-tRNA translocation intermediates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4V6P
 
 | | Structural characterization of mRNA-tRNA translocation intermediates (class 4b of the six classes) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Liao, H, Schreiner, E, Fu, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-12-08 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Structural characterization of mRNA-tRNA translocation intermediates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7YRN
 
 | |
4V8M
 
 | | High-resolution cryo-electron microscopy structure of the Trypanosoma brucei ribosome | | Descriptor: | 18S RRNA OF THE SMALL RIBOSOMAL SUBUNIT, 40S RIBOSOMAL PROTEIN S10, PUTATIVE, ... | | Authors: | Hashem, Y, des Georges, A, Fu, J, Buss, S.N, Jossinet, F, Jobe, A, Zhang, Q, Liao, H.Y, Grassucci, R.A, Bajaj, C, Westhof, E, Madison-Antenucci, S, Frank, J. | | Deposit date: | 2012-12-09 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (5.57 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure of the Trypanosoma Brucei Ribosome.
Nature, 494, 2013
|
|
4V6O
 
 | | Structural characterization of mRNA-tRNA translocation intermediates (class 4a of the six classes) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Liao, H, Schreiner, E, Fu, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structural characterization of mRNA-tRNA translocation intermediates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4V6S
 
 | | Structural characterization of mRNA-tRNA translocation intermediates (class 3 of the six classes) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Liao, H, Schreiner, E, Fu, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-12-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (13.1 Å) | | Cite: | Structural characterization of mRNA-tRNA translocation intermediates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4C4Q
 
 | | Cryo-EM map of the CSFV IRES in complex with the small ribosomal 40S subunit and DHX29 | | Descriptor: | INTERNAL RIBOSOMAL ENTRY SITE | | Authors: | Hashem, Y, desGeorges, A, Dhote, V, Langlois, R, Liao, H.Y, Grassucci, R.A, Pestova, T.V, Hellen, C.U.T, Frank, J. | | Deposit date: | 2013-09-07 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Hepatitis-C-Virus-Like Internal Ribosome Entry Sites Displace Eif3 to Gain Access to the 40S Subunit
Nature, 503, 2013
|
|
3MNV
 
 | | Crystal structure of the non-neutralizing HIV antibody 13H11 Fab fragment | | Descriptor: | 1,2-ETHANEDIOL, ANTI-HIV-1 ANTIBODY 13H11 HEAVY CHAIN, ANTI-HIV-1 ANTIBODY 13H11 LIGHT CHAIN, ... | | Authors: | Nicely, N.I, Dennison, S.M, Kelsoe, G, Liao, H.-X, Alam, S.M, Haynes, B.F. | | Deposit date: | 2010-04-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a non-neutralizing antibody to the HIV-1 gp41 membrane-proximal external region.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4N0T
 
 | | Core structure of the U6 small nuclear ribonucleoprotein at 1.7 Angstrom resolution | | Descriptor: | SULFATE ION, U4/U6 snRNA-associated-splicing factor PRP24, U6 snRNA | | Authors: | Montemayor, E.J, Curran, E.C, Liao, H, Andrews, K.L, Treba, C.N, Butcher, S.E, Brow, D.A. | | Deposit date: | 2013-10-02 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Core structure of the U6 small nuclear ribonucleoprotein at 1.7- angstrom resolution.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4QF1
 
 | | Crystal structure of unliganded CH59UA, the inferred unmutated ancestor of the RV144 anti-HIV antibody lineage producing CH59 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CH59UA Fab fragment of heavy chain, CHLORIDE ION, ... | | Authors: | Wiehe, K, Easterhoff, D, Luo, K, Nicely, N.I, Bradley, T, Jaeger, F.H, Dennison, S.M, Zhang, R, Lloyd, K.E, Stolarchuk, C, Parks, R, Sutherland, L.L, Scearce, R.M, Morris, L, Kaewkungwal, J, Nitayaphan, S, Pitisuttithum, P, Rerks-Ngarm, S, Michael, N, Kim, J, Kelsoe, G, Montefiori, D.C, Tomaras, G, Bonsignori, M, Santra, S, Kepler, T.B, Alam, S.M, Moody, M.A, Liao, H.-X, Haynes, B.F. | | Deposit date: | 2014-05-19 | | Release date: | 2015-02-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antibody Light-Chain-Restricted Recognition of the Site of Immune Pressure in the RV144 HIV-1 Vaccine Trial Is Phylogenetically Conserved.
Immunity, 41, 2014
|
|
4RIS
 
 | | Structural Analysis of the Unmutated Ancestor of the HIV-1 Envelope V2 Region Antibody CH58 Isolated From an RV144 HIV-1 Vaccine Efficacy Trial Vaccinee and Associated with Decreased Transmission Risk | | Descriptor: | CH58-UA Fab heavy chain, CH58-UA Fab light chain, Envelope glycoprotein | | Authors: | Nicely, N.I, Wiehe, K, Kepler, T.B, Jaeger, F.H, Dennison, S.M, Liao, H.-X, Alam, S.M, Hwang, K.-K, Bonsignori, M, Rerks-Ngarm, S, Nitayaphan, S, Pitisuttithum, P, Kaewkungwal, J, Robb, M.L, O'Connell, R.J, Michael, N.L, Kim, J.H, Haynes, B.F. | | Deposit date: | 2014-10-07 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the unmutated ancestor of the HIV-1 envelope V2 region antibody CH58 isolated from an RV144 vaccine efficacy trial vaccinee.
EBioMedicine, 2, 2015
|
|
4RIR
 
 | | Structural Analysis of the Unmutated Ancestor of the HIV-1 Envelope V2 Region Antibody CH58 Isolated From an RV144 HIV-1 Vaccine Efficacy Trial Vaccinee and Associated with Decreased Transmission Risk | | Descriptor: | CH58-UA Fab heavy chain, CH58-UA Fab light chain | | Authors: | Nicely, N.I, Wiehe, K, Kepler, T.B, Jaeger, F.H, Dennison, S.M, Liao, H.-X, Alam, S.M, Hwang, K.-K, Bonsignori, M, Rerks-Ngarm, S, Nitayaphan, S, Pitisuttithum, P, Kaewkungwal, J, Robb, M.L, O'Connell, R.J, Michael, N.L, Kim, J.H, Haynes, B.F. | | Deposit date: | 2014-10-07 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the unmutated ancestor of the HIV-1 envelope V2 region antibody CH58 isolated from an RV144 vaccine efficacy trial vaccinee.
EBioMedicine, 2, 2015
|
|
3MOD
 
 | | Crystal structure of the neutralizing HIV antibody 2F5 Fab fragment (recombinantly produced IgG) with 11 aa gp41 MPER-derived peptide | | Descriptor: | ANTI-HIV-1 ANTIBODY 2F5 HEAVY CHAIN, ANTI-HIV-1 ANTIBODY 2F5 LIGHT CHAIN, gp41 MPER-derived peptide | | Authors: | Nicely, N.I, Dennison, S.M, Kelsoe, G, Liao, H.-X, Alam, S.M, Haynes, B.F. | | Deposit date: | 2010-04-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Non-Neutralizing HIV-1 gp41 Envelope Antibody Demonstrates Neutralization Mechanism of gp41 Antibodies
To be Published
|
|
