2KHS
 
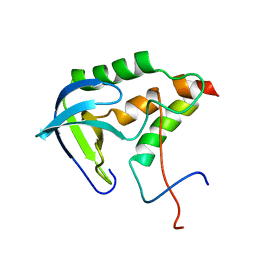 | | Solution structure of SNase121:SNase(111-143) complex | | Descriptor: | Nuclease, Thermonuclease | | Authors: | Geng, Y, Feng, Y, Xie, T, Shan, L, Wang, J. | | Deposit date: | 2009-04-10 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The native-like interactions between SNase121 and SNase(111-143) fragments induce the recovery of their native-like structures and the ability to degrade DNA.
Biochemistry, 48, 2009
|
|
6XEY
 
 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-4 | | Descriptor: | 2-4 Heavy Chain, 2-4 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rapp, M, Shapiro, L, Ho, D.D. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-22 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike.
Nature, 584, 2020
|
|
6XF5
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (RBDs down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
6XF6
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
1I7E
 
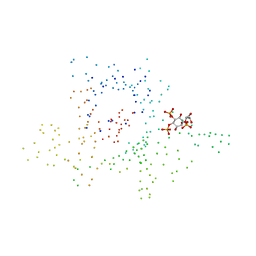 | | C-Terminal Domain Of Mouse Brain Tubby Protein bound to Phosphatidylinositol 4,5-bis-phosphate | | Descriptor: | L-ALPHA-GLYCEROPHOSPHO-D-MYO-INOSITOL-4,5-BIS-PHOSPHATE, TUBBY PROTEIN | | Authors: | Santagata, S, Boggon, T.J, Baird, C.L, Shan, W.S, Shapiro, L. | | Deposit date: | 2001-03-08 | | Release date: | 2001-06-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | G-protein signaling through tubby proteins.
Science, 292, 2001
|
|
8DW9
 
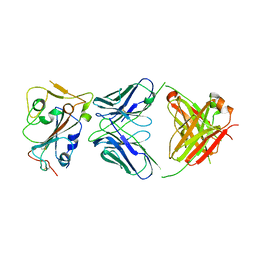 | |
8DXS
 
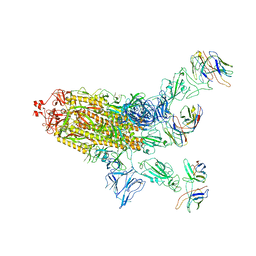 | |
8DWA
 
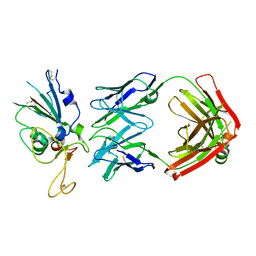 | |
1C8Z
 
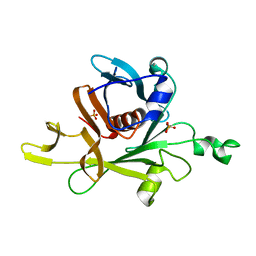 | |
7L5B
 
 | |
7KS9
 
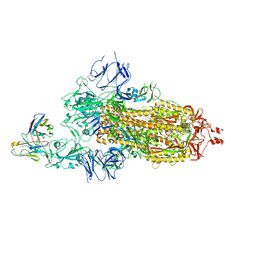 | |
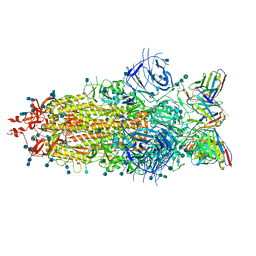
7LS9
 
 | |
7LSS
 
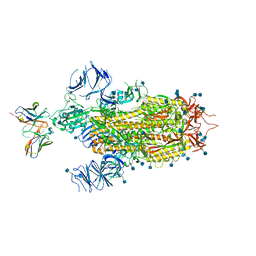 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 2-7 variable heavy chain, ... | | Authors: | Rapp, M, Shapiro, L. | | Deposit date: | 2021-02-18 | | Release date: | 2021-03-17 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural basis for accommodation of emerging B.1.351 and B.1.1.7 variants by two potent SARS-CoV-2 neutralizing antibodies.
Structure, 29, 2021
|
|
7N5H
 
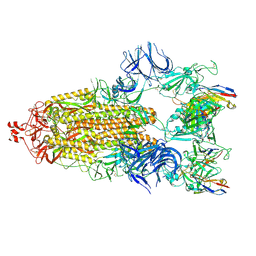 | | Cryo-EM structure of broadly neutralizing antibody 2-36 in complex with prefusion SARS-CoV-2 spike glycoprotein | | Descriptor: | 2-36 Fab heavy chain, 2-36 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Casner, R.G, Cerutti, G, Shapiro, L. | | Deposit date: | 2021-06-05 | | Release date: | 2021-11-03 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | A monoclonal antibody that neutralizes SARS-CoV-2 variants, SARS-CoV, and other sarbecoviruses.
Emerg Microbes Infect, 11, 2022
|
|
4Q7C
 
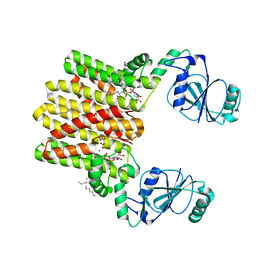 | | Structure of AF2299, a CDP-alcohol phosphotransferase | | Descriptor: | AF2299, a CDP-alcohol phosphotransferase, CALCIUM ION, ... | | Authors: | Clarke, O.B, Sciara, G, Tomasek, D, Banerjee, S, Rajashankar, K.R, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-04-24 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural basis for catalysis in a CDP-alcohol phosphotransferase.
Nat Commun, 5, 2014
|
|
1B2W
 
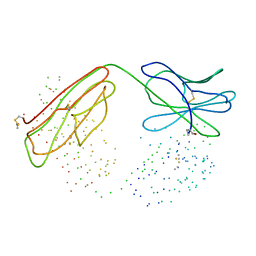 | | COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF A HUMANIZED AND A CHIMERIC FAB OF AN ANTI-GAMMA-INTERFERON ANTIBODY | | Descriptor: | PROTEIN (ANTIBODY (HEAVY CHAIN)), PROTEIN (ANTIBODY (LIGHT CHAIN)) | | Authors: | Fan, Z, Shan, L, Goldsteen, B.Z, Guddat, L.W, Thakur, A, Landolfi, N.F, Co, M.S, Vasquez, M, Queen, C, Ramsland, P.A, Edmundson, A.B. | | Deposit date: | 1998-12-01 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Comparison of the three-dimensional structures of a humanized and a chimeric Fab of an anti-gamma-interferon antibody.
J.Mol.Recog., 12, 1999
|
|
1B4J
 
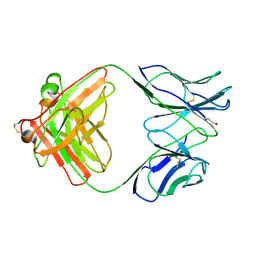 | | COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF A HUMANIZED AND A CHIMERIC FAB OF AN ANTI-GAMMA-INTERFERON ANTIBODY | | Descriptor: | ANTIBODY | | Authors: | Fan, Z, Shan, L, Goldsteen, B.Z, Guddat, L.W, Thakur, A, Landolfi, N.F, Co, M.S, Vasques, M, Queen, C, Ramsland, P.A, Edmundson, A.B. | | Deposit date: | 1998-12-22 | | Release date: | 1999-06-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Comparison of the three-dimensional structures of a humanized and a chimeric Fab of an anti-gamma-interferon antibody.
J.Mol.Recog., 12, 1999
|
|
1AY0
 
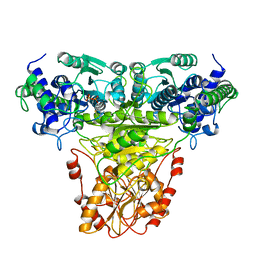 | | IDENTIFICATION OF CATALYTICALLY IMPORTANT RESIDUES IN YEAST TRANSKETOLASE | | Descriptor: | CALCIUM ION, THIAMINE DIPHOSPHATE, TRANSKETOLASE | | Authors: | Wikner, C, Nilsson, U, Meshalkina, L, Udekwu, C, Lindqvist, Y, Schneider, G. | | Deposit date: | 1997-11-13 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of catalytically important residues in yeast transketolase.
Biochemistry, 36, 1997
|
|
8G4M
 
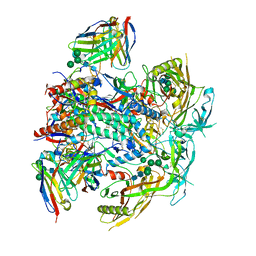 | | Vaccine-elicited human antibody 2C06 in complex with HIV-1 envelope trimer BG505 DS-SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Wang, S, Morano, N.C, Shapiro, L, Kwong, P.D. | | Deposit date: | 2023-02-10 | | Release date: | 2023-07-12 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | HIV-1 neutralizing antibodies elicited in humans by a prefusion-stabilized envelope trimer form a reproducible class targeting fusion peptide.
Cell Rep, 42, 2023
|
|
1ETZ
 
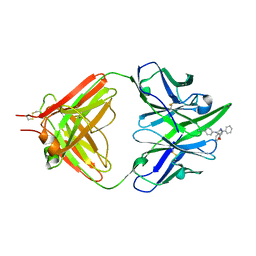 | | THE THREE-DIMENSIONAL STRUCTURE OF AN ANTI-SWEETENER FAB, NC10.14, SHOWS THE EXTENT OF STRUCTURAL DIVERSITY IN ANTIGEN RECOGNITION BY IMMUNOGLOBULINS | | Descriptor: | FAB NC10.14 - HEAVY CHAIN, FAB NC10.14 - LIGHT CHAIN, N-(P-CYANOPHENYL)-N'-DIPHENYLMETHYL-GUANIDINE-ACETIC ACID | | Authors: | Guddat, L.W, Shan, L, Broomell, C, Ramsland, P.A, Fan, Z, Anchin, J.M, Linthicum, D.S, Edmundson, A.B. | | Deposit date: | 2000-04-13 | | Release date: | 2000-10-18 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The three-dimensional structure of a complex of a murine Fab (NC10. 14) with a potent sweetener (NC174): an illustration of structural diversity in antigen recognition by immunoglobulins.
J.Mol.Biol., 302, 2000
|
|
1NCJ
 
 | | N-CADHERIN, TWO-DOMAIN FRAGMENT | | Descriptor: | CALCIUM ION, PROTEIN (N-CADHERIN), URANYL (VI) ION | | Authors: | Tamura, K, Shan, W.-S, Hendrickson, W.A, Colman, D.R, Shapiro, L. | | Deposit date: | 1999-02-02 | | Release date: | 1999-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure-function analysis of cell adhesion by neural (N-) cadherin.
Neuron, 20, 1998
|
|
8DEQ
 
 | | Cryo-EM local refinement of antibody SKV09 in complex with VEEV alphavirus spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SKV09 Fab Heavy Chain, SKV09 Fab Light Chain, ... | | Authors: | Casner, R.G, Verardi, R, Roederer, M, Shapiro, L. | | Deposit date: | 2022-06-21 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
8DER
 
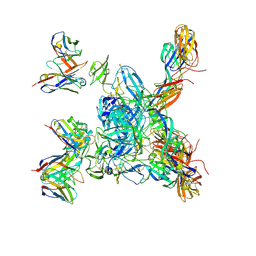 | | Cryo-EM local refinement of antibody SKV16 in complex with VEEV alphavirus spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SKV16 Fab Heavy Chain, ... | | Authors: | Casner, R.G, Verardi, R, Roederer, M, Shapiro, L. | | Deposit date: | 2022-06-21 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
3LNE
 
 | |
3LNF
 
 | |
