3JSX
 
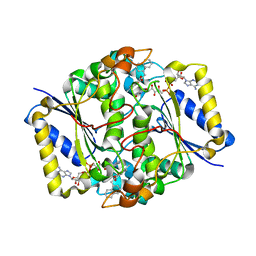 | | X-ray Crystal structure of NAD(P)H: Quinone Oxidoreductase-1 (NQO1) bound to the coumarin-based inhibitor AS1 | | Descriptor: | 4-hydroxy-6,7-dimethyl-3-(naphthalen-1-ylmethyl)-2H-chromen-2-one, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Dunstan, M.S, Levy, C, Leys, D. | | Deposit date: | 2009-09-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Synthesis and biological evaluation of coumarin-based inhibitors of NAD(P)H: quinone oxidoreductase-1 (NQO1).
J.Med.Chem., 52, 2009
|
|
4ASG
 
 | | The structure of modified benzoquinone ansamycins bound to yeast N- terminal Hsp90 | | Descriptor: | ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, NICKEL (II) ION, [(3R,5S,6R,7R,11S,12Z,14E)-5,11,21-trimethoxy-3,7,9,15-tetramethyl-6-oxidanyl-16,20,22-tris(oxidanylidene)-19-phenyl-17-azabicyclo[16.3.1]docosa-1(21),8,12,14,18-pentaen-10-yl] carbamate | | Authors: | Roe, S.M, Prodromou, C. | | Deposit date: | 2012-05-01 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis of 19-Substituted Geldanamycins with Altered Conformations and Their Binding to Heat Shock Protein Hsp90.
Nat.Chem., 5, 2013
|
|
4AS9
 
 | | The structure of modified benzoquinone ansamycins bound to yeast N- terminal Hsp90 | | Descriptor: | ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, [(3R,5S,6R,7R,10R,11S,12E)-5,11,21-trimethoxy-3,7,9,15,19-pentamethyl-6-oxidanyl-16,20,22-tris(oxidanylidene)-17-azabicyclo[16.3.1]docosa-1(21),8,12,14,18-pentaen-10-yl] carbamate | | Authors: | Roe, S.M, Prodromou, C. | | Deposit date: | 2012-04-30 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Synthesis of 19-substituted geldanamycins with altered conformations and their binding to heat shock protein Hsp90.
Nat Chem, 5, 2013
|
|
4ASF
 
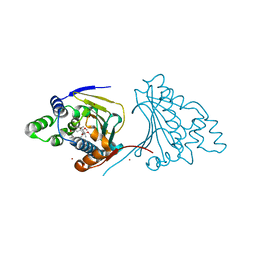 | | The structure of modified benzoquinone ansamycins bound to yeast N- terminal Hsp90 | | Descriptor: | (8S,9R,13R,14S,16R)-21-(furan-2-yl)-13-hydroxy-8,14,19-trimethoxy-16-methyl-4,10,12-trimethylidene-3,20,22-trioxo-2-azabicyclo[16.3.1]docosa-1(21),18-dien-9-yl carbamate, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, NICKEL (II) ION | | Authors: | Roe, S.M, Prodromou, C. | | Deposit date: | 2012-05-01 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis of 19-substituted geldanamycins with altered conformations and their binding to heat shock protein Hsp90.
Nat Chem, 5, 2013
|
|
4ASA
 
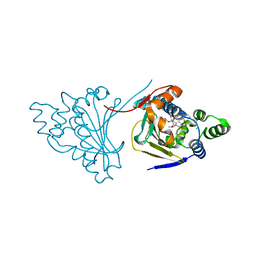 | | The structure of modified benzoquinone ansamycins bound to yeast N- terminal Hsp90 | | Descriptor: | ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, CHLORIDE ION, [(3R,5S,6R,7R,12E)-5,11-dimethoxy-3,7,9,15,19-pentamethyl-6-oxidanyl-16,20,22-tris(oxidanylidene)-21-(prop-2-enylamino)-17-azabicyclo[16.3.1]docosa-1(21),8,12,14,18-pentaen-10-yl] carbamate | | Authors: | Roe, S.M, Prodromou, C. | | Deposit date: | 2012-04-30 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Synthesis of 19-Substituted Geldanamycins with Altered Conformations and Their Binding to Heat Shock Protein Hsp90.
Nat.Chem., 5, 2013
|
|
4ASB
 
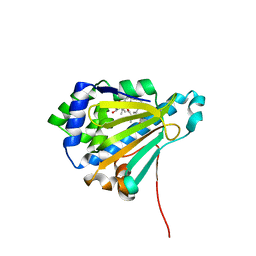 | | The structure of modified benzoquinone ansamycins bound to yeast N- terminal Hsp90 | | Descriptor: | (4E,6E,8S,9R,10E,12R,13R,14S,16R)-19-{[2-(dimethylamino)ethyl]amino}-13-hydroxy-8,14-dimethoxy-4,10,12,16,21-pentamethyl-3,20,22-trioxo-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Roe, S.M, Prodromou, C. | | Deposit date: | 2012-04-30 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Synthesis of 19-substituted geldanamycins with altered conformations and their binding to heat shock protein Hsp90.
Nat Chem, 5, 2013
|
|
5KGN
 
 | | 1.95A resolution structure of independent phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (2d) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
5KGM
 
 | | 2.95A resolution structure of Apo independent phosphoglycerate mutase from C. elegans (monoclinic form) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
5KGL
 
 | | 2.45A resolution structure of Apo independent phosphoglycerate mutase from C. elegans (orthorhombic form) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
7KC4
 
 | | Human WLS in complex with WNT8A | | Descriptor: | 1-CIS-9-OCTADECANOYL-2-CIS-9-HEXADECANOYL PHOSPHATIDYL GLYCEROL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Nygaard, R, Jia, Y, Kim, J, Ross, D, Parisi, G, Clarke, O.B, Virshup, D.M, Mancia, F. | | Deposit date: | 2020-10-05 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural Basis of WLS/Evi-Mediated Wnt Transport and Secretion.
Cell, 184, 2021
|
|
1ETT
 
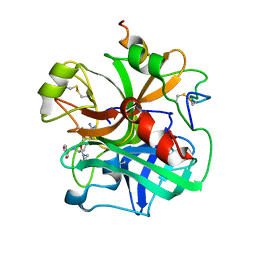 | |
2VX0
 
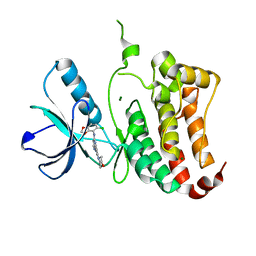 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION, N'-(5-CHLORO-1,3-BENZODIOXOL-4-YL)-N-(3-MORPHOLIN-4-YLPHENYL)PYRIMIDINE-2,4-DIAMINE | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Leach, A.G, Kettle, J.G. | | Deposit date: | 2008-06-30 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 1: Structure-Based Design and Optimization of a Series of 2,4-Bis-Anilinopyrimidines.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VWY
 
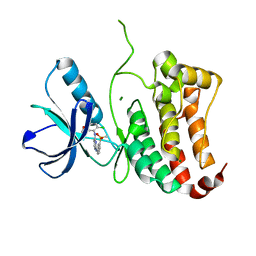 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION, N'-(5-chloro-1,3-benzodioxol-4-yl)-N-(3-methylsulfonylphenyl)pyrimidine-2,4-diamine | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Leach, A.G, Kettle, J.G. | | Deposit date: | 2008-06-30 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 2: Structure-Based Discovery and Optimisation of 3,5-Bis Substituted Anilinopyrimidines.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VWZ
 
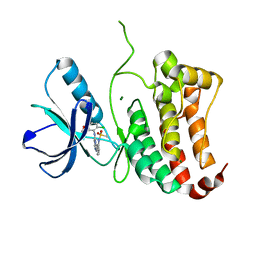 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION, N-[3-[[4-[(5-CHLORO-1,3-BENZODIOXOL-4-YL)AMINO]PYRIMIDIN-2-YL]AMINO]PHENYL]METHANESULFONAMIDE | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Leach, A.G, Kettle, J.G. | | Deposit date: | 2008-06-30 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 2: Structure-Based Discovery and Optimisation of 3,5-Bis Substituted Anilinopyrimidines.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VX1
 
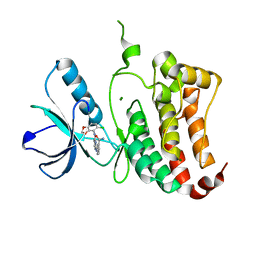 | | ephB4 kinase domain inhibitor complex | | Descriptor: | 3-({4-[(5-CHLORO-1,3-BENZODIOXOL-4-YL)AMINO]PYRIMIDIN-2-YL}AMINO)BENZAMIDE, EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Leach, A.G, Kettle, J.G. | | Deposit date: | 2008-06-30 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 2: Structure-Based Discovery and Optimisation of 3,5-Bis Substituted Anilinopyrimidines.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VWX
 
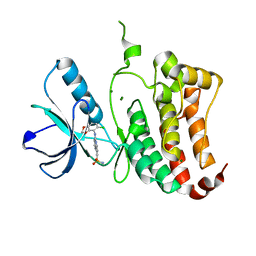 | | ephB4 kinase domain inhibitor complex | | Descriptor: | 3-({4-[(5-chloro-1,3-benzodioxol-4-yl)amino]pyrimidin-2-yl}amino)benzenesulfonamide, EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Leach, A.G, Kettle, J.G. | | Deposit date: | 2008-06-27 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 2: Structure-Based Discovery and Optimisation of 3,5-Bis Substituted Anilinopyrimidines.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1ETR
 
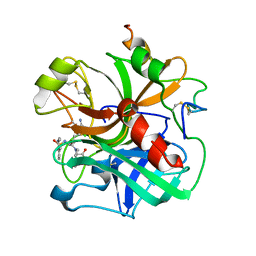 | | REFINED 2.3 ANGSTROMS X-RAY CRYSTAL STRUCTURE OF BOVINE THROMBIN COMPLEXES FORMED WITH THE BENZAMIDINE AND ARGININE-BASED THROMBIN INHIBITORS NAPAP, 4-TAPAP AND MQPA: A STARTING POINT FOR IMPROVING ANTITHROMBOTICS | | Descriptor: | EPSILON-THROMBIN, amino{[(4S)-5-[(2R,4R)-2-carboxy-4-methylpiperidin-1-yl]-4-({[(3R)-3-methyl-1,2,3,4-tetrahydroquinolin-8-yl]sulfonyl}amino)-5-oxopentyl]amino}methaniminium | | Authors: | Bode, W, Brandstetter, H. | | Deposit date: | 1992-07-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined 2.3 A X-ray crystal structure of bovine thrombin complexes formed with the benzamidine and arginine-based thrombin inhibitors NAPAP, 4-TAPAP and MQPA. A starting point for improving antithrombotics.
J.Mol.Biol., 226, 1992
|
|
1ETS
 
 | |
9C1F
 
 | |
9C1E
 
 | | Mink RyR3 in closed conformation | | Descriptor: | CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 3, ... | | Authors: | Chen, Y.S, Van Petegem, F. | | Deposit date: | 2024-05-29 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM investigation of ryanodine receptor type 3.
Nat Commun, 15, 2024
|
|
2X9F
 
 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION, N^4^-1H-INDAZOL-4-YL-N^2^-[3-(METHYLSULFONYL)PHENYL]PYRIMIDINE-2,4-DIAMINE | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D. | | Deposit date: | 2010-03-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 3: Identification of Non-Benzodioxole-Based Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4LI5
 
 | | EGFR-K IN COMPLEX WITH N-[3-[[5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]-4-methoxy-phenyl] Prop-2-enamide | | Descriptor: | Epidermal growth factor receptor, N-(3-{[5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl]amino}-4-methoxyphenyl)propanamide, SODIUM ION | | Authors: | Debreczeni, J.E, Seiffert, G.B, Kiefersauer, R, Augustin, M, Nagel, S, Ward, R, Anderton, M, Ashton, S, Bethel, P, Box, M, Butterworth, S, Colclough, N, Chroley, C, Chuaqui, C, Cross, D, Eberlein, C, Finlay, R, Hill, G, Grist, M, Klinowska, T, Lane, C, Martin, S, Orme, J, Smith, P, Wang, F, Waring, M. | | Deposit date: | 2013-07-02 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure- and Reactivity-Based Development of Covalent Inhibitors of the Activating and Gatekeeper Mutant Forms of the Epidermal Growth Factor Receptor (EGFR).
J.Med.Chem., 56, 2013
|
|
2VWU
 
 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, N-(5-chloro-1,3-benzodioxol-4-yl)-6-methoxy-7-(3-piperidin-1-ylpropoxy)quinazolin-4-amine | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Rowsell, S, Packer, M, McAlister, M. | | Deposit date: | 2008-06-27 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 1: Structure-Based Design and Optimization of a Series of 2,4-Bis-Anilinopyrimidines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VWW
 
 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, N'-(5-CHLORO-1,3-BENZODIOXOL-4-YL)-N-(3,4,5- TRIMETHOXYPHENYL)PYRIMIDINE-2,4-DIAMINE | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Leach, A.G, Kettle, J.G. | | Deposit date: | 2008-06-27 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 1: Structure-Based Design and Optimization of a Series of 2,4-Bis-Anilinopyrimidines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1PPB
 
 | |
