6IYU
 
 | | Crystal Structure Analysis of an Eukaryotic Membrane Protein | | Descriptor: | CALCIUM ION, CHLORIDE ION, Trimeric intracellular cation channel type A | | Authors: | Wang, X.H, Zeng, Y, Su, M, Gao, F, Chen, Y.H. | | Deposit date: | 2018-12-17 | | Release date: | 2019-05-01 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IZ1
 
 | | Crystal Structure Analysis of a Eukaryotic Membrane Protein | | Descriptor: | CALCIUM ION, CHLORIDE ION, Trimeric intracellular cation channel type A | | Authors: | Wang, X.H, Zeng, Y, Gao, F, Su, M, Chen, Y.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IZ0
 
 | | Crystal Structure Analysis of a Eukaryotic Membrane Protein | | Descriptor: | CALCIUM ION, CHLORIDE ION, Trimeric intracellular cation channel type A | | Authors: | Wang, X.H, Zeng, Y, Gao, F, Su, M, Chen, Y.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IZ6
 
 | | Crystal Structure Analysis of TRIC counter-ion channels in calcium release | | Descriptor: | CALCIUM ION, Trimeric intracellular cation channel type B-B | | Authors: | Liu, X.L, Wang, X.H, Su, M, Hendrickson, W.A, Chen, Y.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.293 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IZF
 
 | | Structural basis for activity of TRIC counter-ion channels in calcium release | | Descriptor: | (2R)-1-(dodecanoyloxy)-3-hydroxypropan-2-yl (5E,8E,11E)-tetradeca-5,8,11-trienoate, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, X.H, Zeng, Y, Su, M, Hendrickson, W.A, Chen, Y.H. | | Deposit date: | 2018-12-19 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IZ3
 
 | |
7F46
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab (state1, local refinement of the RBD, NTD and 35B5 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-06-17 | | Release date: | 2022-03-23 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (4.79 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
5YTK
 
 | | Crystal structure of SIRT3 bound to a leucylated AceCS2 | | Descriptor: | AceCS2-KLeu, LEUCINE, LYSINE, ... | | Authors: | Li, J, Gong, W, Xu, Y. | | Deposit date: | 2017-11-18 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sensing and Transmitting Intracellular Amino Acid Signals through Reversible Lysine Aminoacylations
Cell Metab., 27, 2018
|
|
5YBJ
 
 | | Structure of apo KANK1 ankyrin domain | | Descriptor: | GLYCEROL, KN motif and ankyrin repeat domain-containing protein 1 | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
5YBV
 
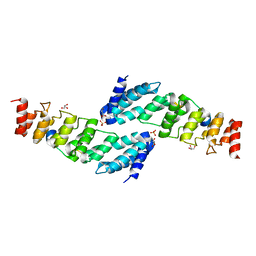 | | The structure of the KANK2 ankyrin domain with the KIF21A peptide | | Descriptor: | GLYCEROL, KN motif and ankyrin repeat domain-containing protein 2, Kinesin-like protein KIF21A, ... | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
5YBU
 
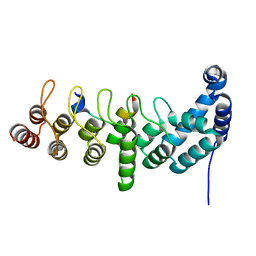 | | Structure of the KANK1 ankyrin domain in complex with KIF21A peptide | | Descriptor: | KN motif and ankyrin repeat domain-containing protein 1, Kinesin-like protein KIF21A | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
6IUP
 
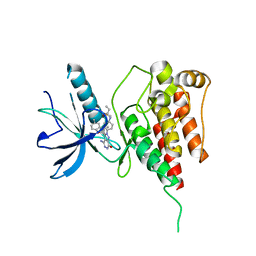 | | Crystal structure of FGFR4 kinase domain in complex with compound 5 | | Descriptor: | DIMETHYL SULFOXIDE, Fibroblast growth factor receptor 4, N-{4-[4-amino-3-(3,5-dimethyl-1-benzofuran-2-yl)-7-oxo-6,7-dihydro-2H-pyrazolo[3,4-d]pyridazin-2-yl]phenyl}prop-2-enamide | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-29 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
6IYF
 
 | | Structure of pSTING complex | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-12-15 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
7ESI
 
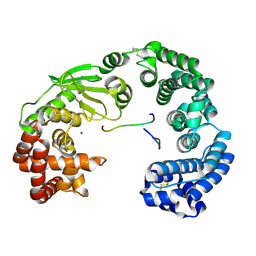 | | Crystal structure of the collagenase unit of a Vibrio collagenase from Vibrio harveyi VHJR7 at 1. 8 angstrom resolution. | | Descriptor: | CALCIUM ION, Collagenase unit (CU), Peptide P1, ... | | Authors: | Cao, H.Y, Wang, Y, Peng, M, Zhang, Y.Z. | | Deposit date: | 2021-05-11 | | Release date: | 2022-02-09 | | Last modified: | 2023-02-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Vibrio collagenase VhaC provides insight into the mechanism of bacterial collagenolysis.
Nat Commun, 13, 2022
|
|
4QQN
 
 | | Protein arginine methyltransferase 3 in complex with compound MTV044246 | | Descriptor: | 1-{2-[1-(aminomethyl)cyclohexyl]ethyl}-3-isoquinolin-6-ylurea, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dong, A, Dobrovetsky, E, Tempel, W, He, H, Zhao, K, Smil, D, Landon, M, Luo, X, Chen, Z, Dai, M, Yu, Z, Lin, Y, Zhang, H, Zhao, K, Schapira, M, Brown, P.J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of Potent and Selective Allosteric Inhibitors of Protein Arginine Methyltransferase 3 (PRMT3).
J. Med. Chem., 61, 2018
|
|
7EVR
 
 | | Crystal structure of hnRNP L RRM2 in complex with SETD2 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein L, SHI domain from Histone-lysine N-methyltransferase SETD2 | | Authors: | Li, F.D, Wang, S.M. | | Deposit date: | 2021-05-22 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the interaction between SETD2 methyltransferase and hnRNP L paralogs for governing co-transcriptional splicing.
Nat Commun, 12, 2021
|
|
7EVS
 
 | | Crystal structure of hnRNP LL RRM2 in complex with SETD2 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein L-like, SHI domain from Histone-lysine N-methyltransferase SETD2, SULFATE ION | | Authors: | Li, F.D, Wang, S.M. | | Deposit date: | 2021-05-22 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of the interaction between SETD2 methyltransferase and hnRNP L paralogs for governing co-transcriptional splicing.
Nat Commun, 12, 2021
|
|
7KDT
 
 | |
6P7I
 
 | | Crystal structure of Human PRMT6 in complex with S-Adenosyl-L-Homocysteine and YS17-117 Compound | | Descriptor: | GLYCEROL, N-[3-(4-{[(2-aminoethyl)(methyl)amino]methyl}-1H-pyrrol-3-yl)phenyl]prop-2-enamide, N-[3-(4-{[(2-aminoethyl)(methyl)amino]methyl}-1H-pyrrol-3-yl)phenyl]propanamide, ... | | Authors: | Halabelian, L, Dong, A, Zeng, H, Li, Y, Seitova, A, Hutchinson, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-05 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a First-in-Class Protein Arginine Methyltransferase 6 (PRMT6) Covalent Inhibitor
J.Med.Chem., 63, 2020
|
|
7F75
 
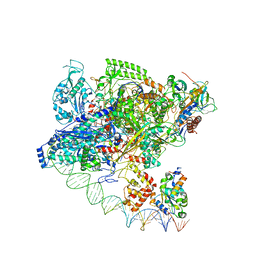 | | Cryo-EM structure of Spx-dependent transcription activation complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Feng, Y, Shi, J. | | Deposit date: | 2021-06-28 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of transcription activation by the global regulator Spx.
Nucleic Acids Res., 49, 2021
|
|
5Y5P
 
 | |
5Y5Q
 
 | |
5YJ8
 
 | |
5Y5O
 
 | |
5XVA
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH216 | | Descriptor: | ETHANOL, Serine/threonine-protein kinase PAK 4, [6-chloranyl-4-[(5-methyl-1H-pyrazol-3-yl)amino]quinazolin-2-yl]-[(3R)-3-methylpiperazin-1-yl]methanone | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
