5LUS
 
 | | Structures of DHBN domain of Pelecanus crispus BLM helicase | | Descriptor: | BLM helicase | | Authors: | Shi, J, Chen, W.-F, Zhang, B, Fan, S.-H, Ai, X, Liu, N.-N, Rety, S, Xi, X.-G. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-01 | | Last modified: | 2017-04-19 | | Method: | X-RAY DIFFRACTION (1.433 Å) | | Cite: | A helical bundle in the N-terminal domain of the BLM helicase mediates dimer and potentially hexamer formation.
J. Biol. Chem., 292, 2017
|
|
5LUT
 
 | | Structures of DHBN domain of Gallus gallus BLM helicase | | Descriptor: | BLM helicase, PHOSPHATE ION | | Authors: | Shi, J, Chen, W.-F, Zhang, B, Fan, S.-H, Ai, X, Liu, N.-N, Rety, S, Xi, X.-G. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-01 | | Last modified: | 2017-04-19 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A helical bundle in the N-terminal domain of the BLM helicase mediates dimer and potentially hexamer formation.
J. Biol. Chem., 292, 2017
|
|
5LUP
 
 | | Structures of DHBN domain of human BLM helicase | | Descriptor: | BLM protein, PHOSPHATE ION, POTASSIUM ION | | Authors: | Shi, J, Chen, W.-F, Zhang, B, Fan, S.-H, Ai, X, Liu, N.-N, Rety, S, Xi, X.-G. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-01 | | Last modified: | 2017-04-19 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | A helical bundle in the N-terminal domain of the BLM helicase mediates dimer and potentially hexamer formation.
J. Biol. Chem., 292, 2017
|
|
5MK5
 
 | | Structures of DHBN domain of human BLM helicase | | Descriptor: | Bloom syndrome protein, IODIDE ION, POTASSIUM ION | | Authors: | Shi, J, Chen, W.-F, Zhang, B, Fan, S.-H, Ai, X, Liu, N.-N, Rety, S, Xi, X.-G. | | Deposit date: | 2016-12-02 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A helical bundle in the N-terminal domain of the BLM helicase mediates dimer and potentially hexamer formation.
J. Biol. Chem., 292, 2017
|
|
3IKK
 
 | | Crystal structure analysis of msp domain | | Descriptor: | Vesicle-associated membrane protein-associated protein B/C | | Authors: | Shi, J, Lua, S, Song, J. | | Deposit date: | 2009-08-06 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Elimination of the native structure and solubility of the hVAPB MSP domain by the Pro56Ser mutation that causes amyotrophic lateral sclerosis.
Biochemistry, 49, 2010
|
|
5GNG
 
 | |
5H3B
 
 | |
6M6B
 
 | | Cryo-EM structure of Thermus thermophilus Mfd in complex with RNA polymerase and ATP-gamma-S | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
6M6A
 
 | | Cryo-EM structure of Thermus thermophilus Mfd in complex with RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
6M6C
 
 | | CryoEM structure of Thermus thermophilus RNA polymerase elongation complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
6J31
 
 | | Crystal Structure Analysis of the Glycotransferase of kitacinnamycin | | Descriptor: | (2E,2'E)-3,3'-(1,2-phenylene)di(prop-2-enoic acid), DBB-DSG-VAL-MEA-VAL-GLY-GLY-DVA-DLE, kcn28 | | Authors: | Shi, J, Liu, C.L, Zhang, B, Guo, W.J, Zhu, J.P, Xu, X, Xu, Q, Jiao, R.H, Tan, R.X, Ge, H.M. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Genome mining and biosynthesis of kitacinnamycins as a STING activator.
Chem Sci, 10, 2019
|
|
6J32
 
 | | Crystal Structure Analysis of the Glycotransferase of kitacinnamycin | | Descriptor: | Kcn28 | | Authors: | Shi, J, Liu, C.L, Zhang, B, Guo, W.J, Zhu, J.P, Xu, X, Xu, Q, Jiao, R.H, Tan, R.X, Ge, H.M. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Genome mining and biosynthesis of kitacinnamycins as a STING activator.
Chem Sci, 10, 2019
|
|
7D7D
 
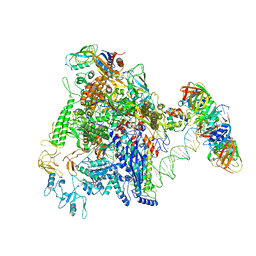 | | CryoEM structure of gp45-dependent transcription activation complex | | Descriptor: | DNA (nontemplate strand), DNA (template strand), DNA polymerase clamp, ... | | Authors: | Shi, J, Wen, A, Jin, S, Feng, Y. | | Deposit date: | 2020-10-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Transcription activation by a sliding clamp.
Nat Commun, 12, 2021
|
|
7D7C
 
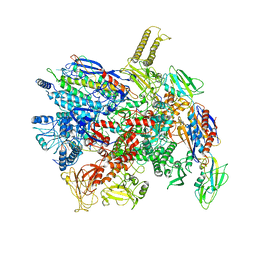 | | CryoEM structure of gp55-dependent RNA polymerase-promoter open complex | | Descriptor: | DNA (nontemplate strand), DNA (template strand), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Shi, J, Wen, A, Jin, S, Feng, Y. | | Deposit date: | 2020-10-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Transcription activation by a sliding clamp.
Nat Commun, 12, 2021
|
|
6K4Y
 
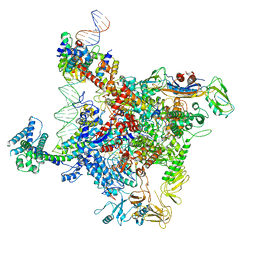 | | CryoEM structure of sigma appropriation complex | | Descriptor: | 10 kDa anti-sigma factor, DNA (60-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structural basis of sigma appropriation.
Nucleic Acids Res., 47, 2019
|
|
2WNJ
 
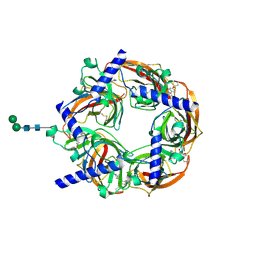 | | CRYSTAL STRUCTURE OF APLYSIA ACHBP IN COMPLEX WITH DMXBA | | Descriptor: | (3E)-3-[(2,4-DIMETHOXYPHENYL)METHYLIDENE]-3,4,5,6-TETRAHYDRO-2,3'-BIPYRIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, SOLUBLE ACETYLCHOLINE RECEPTOR, ... | | Authors: | Sulzenbacher, G, Hibbs, R, Shi, J, Talley, T, Conrod, S, Kem, W, Taylor, P, Marchot, P, Bourne, Y. | | Deposit date: | 2009-07-09 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Determinants for Interaction of Partial Agonists with Acetylcholine Binding Protein and Neuronal Alpha7 Nicotinic Acetylcholine Receptor.
Embo J., 28, 2009
|
|
2WN9
 
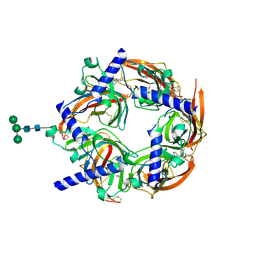 | | Crystal structure of Aplysia ACHBP in complex with 4-0H-DMXBA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(E)-5,6-DIHYDRO-2,3'-BIPYRIDIN-3(4H)-YLIDENEMETHYL]-3-METHOXYPHENOL, SOLUBLE ACETYLCHOLINE RECEPTOR, ... | | Authors: | Sulzenbacher, G, Hibbs, R, Shi, J, Talley, T, Conrod, S, Kem, W, Taylor, P, Marchot, P, Bourne, Y. | | Deposit date: | 2009-07-07 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural determinants for interaction of partial agonists with acetylcholine binding protein and neuronal alpha7 nicotinic acetylcholine receptor.
Embo J., 28, 2009
|
|
2WNL
 
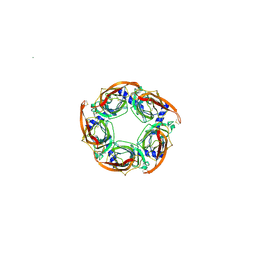 | | CRYSTAL STRUCTURE OF APLYSIA ACHBP IN COMPLEX WITH ANABASEINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,4,5,6-tetrahydro-2,3'-bipyridine, 5-amino-1-pyridin-3-ylpentan-1-one, ... | | Authors: | Sulzenbacher, G, Hibbs, R, Shi, J, Talley, T, Conrod, S, Kem, W, Taylor, P, Marchot, P, Bourne, Y. | | Deposit date: | 2009-07-09 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural determinants for interaction of partial agonists with acetylcholine binding protein and neuronal alpha7 nicotinic acetylcholine receptor.
EMBO J., 28, 2009
|
|
2WNC
 
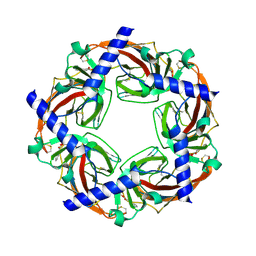 | | Crystal structure of Aplysia ACHBP in complex with tropisetron | | Descriptor: | (3-ENDO)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL 1H-INDOLE-3-CARBOXYLATE, Soluble acetylcholine receptor | | Authors: | Sulzenbacher, G, Hibbs, R, Shi, J, Talley, T, Conrod, S, Kem, W, Taylor, P, Marchot, P, Bourne, Y. | | Deposit date: | 2009-07-08 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural determinants for interaction of partial agonists with acetylcholine binding protein and neuronal alpha7 nicotinic acetylcholine receptor.
EMBO J., 28, 2009
|
|
8HAE
 
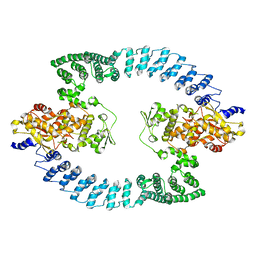 | | Cryo-EM structure of HACE1 dimer | | Descriptor: | E3 ubiquitin-protein ligase HACE1 | | Authors: | Singh, S, Machida, S, Tulsian, N.K, Choong, Y.K, Ng, J, Shanker, S, Yaochen, L.D, Shi, J, Sivaraman, J, Machida, S. | | Deposit date: | 2022-10-26 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structural Basis for the Enzymatic Activity of the HACE1 HECT-Type E3 Ligase Through N-Terminal Helix Dimerization.
Adv Sci, 10, 2023
|
|
8H8X
 
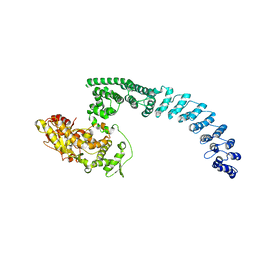 | | Cryo-EM structure of HACE1 monomer | | Descriptor: | E3 ubiquitin-protein ligase HACE1 | | Authors: | Singh, S, Machida, S, Tulsian, N.K, Choong, Y.K, Ng, J, Shanker, S, Yaochen, L.D, Shi, J, Sivaraman, J. | | Deposit date: | 2022-10-24 | | Release date: | 2023-06-28 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structural Basis for the Enzymatic Activity of the HACE1 HECT-Type E3 Ligase Through N-Terminal Helix Dimerization.
Adv Sci, 10, 2023
|
|
1TM9
 
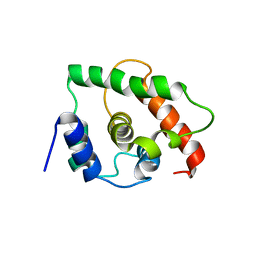 | | NMR Structure of gene target number gi3844938 from Mycoplasma genitalium: Berkeley Structural Genomics Center | | Descriptor: | Hypothetical protein MG354 | | Authors: | Pelton, J.G, Shi, J, Yokota, H, Kim, R, Wemmer, D.E, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-06-10 | | Release date: | 2004-08-10 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Gene Target gi3844938 from Mycoplasma genitalium
To be Published
|
|
8HIH
 
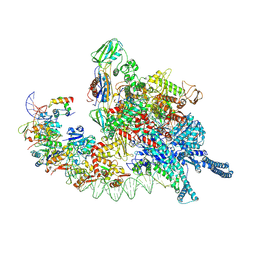 | | Cryo-EM structure of Mycobacterium tuberculosis transcription initiation complex with transcription factor GlnR | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Shi, J, Xu, J.C. | | Deposit date: | 2022-11-20 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural insights into the transcription activation mechanism of the global regulator GlnR from actinobacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5CB2
 
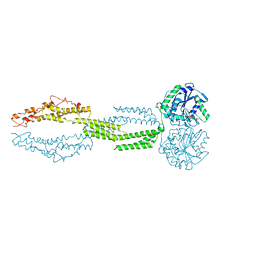 | | the structure of candida albicans Sey1p in complex with GMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Protein SEY1 | | Authors: | Yan, L, Sun, S, Wang, W, Shi, J, Hu, X, Wang, S, Rao, Z, Hu, J, Lou, Z. | | Deposit date: | 2015-06-30 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the yeast dynamin-like GTPase Sey1p provide insight into homotypic ER fusion
J.Cell Biol., 210, 2015
|
|
7VWZ
 
 | | Cryo-EM structure of Rob-dependent transcription activation complex in a unique conformation | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Feng, Y, Shi, J. | | Deposit date: | 2021-11-12 | | Release date: | 2022-06-08 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of transcription activation by Rob, a pleiotropic AraC/XylS family regulator.
Nucleic Acids Res., 50, 2022
|
|
