3AMS
 
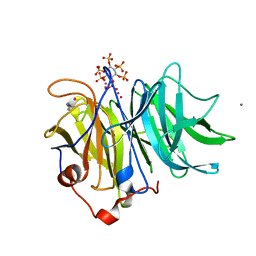 | | Crystal Structures of Bacillus subtilis Alkaline Phytase in Complex with Ca2+, Cd2+, Co2+, Ni2+, Mg2+ and myo-Inositol Hexasulfate | | Descriptor: | 3-phytase, CADMIUM ION, CALCIUM ION, ... | | Authors: | Zeng, Y.F, Ko, T.P, Lai, H.L, Cheng, Y.S, Wu, T.H, Ma, Y, Yang, C.S, Cheng, K.J, Huang, C.H, Guo, R.T, Liu, J.R. | | Deposit date: | 2010-08-22 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structures of Bacillus alkaline phytase in complex with divalent metal ions and inositol hexasulfate
J.Mol.Biol., 409, 2011
|
|
3AMR
 
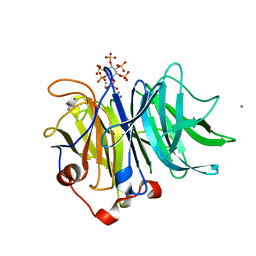 | | Crystal Structures of Bacillus subtilis Alkaline Phytase in Complex with Ca2+, Co2+, Ni2+, Mg2+ and myo-Inositol Hexasulfate | | Descriptor: | 3-phytase, CALCIUM ION, D-MYO-INOSITOL-HEXASULPHATE | | Authors: | Zeng, Y.F, Ko, T.P, Lai, H.L, Cheng, Y.S, Wu, T.H, Ma, Y, Yang, C.S, Cheng, K.J, Huang, C.H, Guo, R.T, Liu, J.R. | | Deposit date: | 2010-08-22 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structures of Bacillus alkaline phytase in complex with divalent metal ions and inositol hexasulfate
J.Mol.Biol., 409, 2011
|
|
6XHJ
 
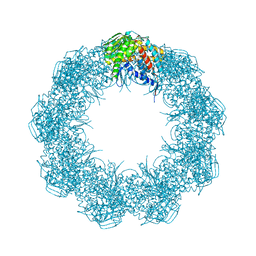 | | Cryo-EM structure of octadecameric TF55 (beta-only) complex from S. solfataricus bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Thermosome subunit beta | | Authors: | Zeng, Y.C, Sobti, M, Stewart, A.G. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural analysis of the Sulfolobus solfataricus TF55beta chaperonin by cryo-electron microscopy
Acta Crystallogr.,Sect.F, 77, 2021
|
|
6XHI
 
 | |
8K5P
 
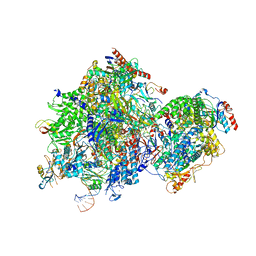 | |
8JRC
 
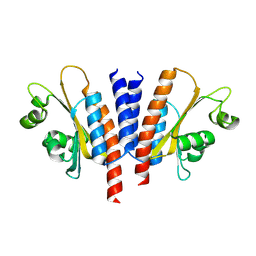 | |
8JCH
 
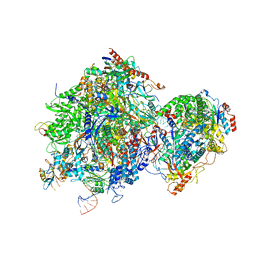 | |
8SC3
 
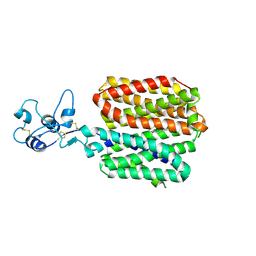 | |
8SC4
 
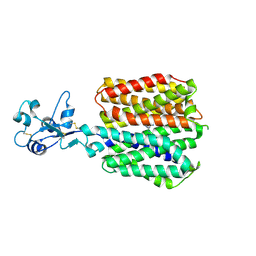 | |
8SC1
 
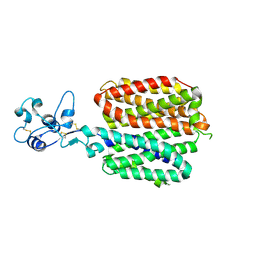 | |
8SC2
 
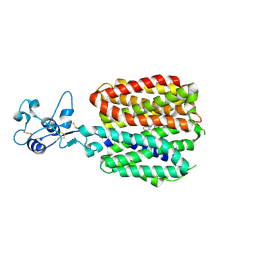 | |
8SC6
 
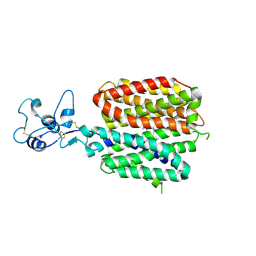 | | Human OCT1 bound to thiamine in inward-open conformation | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, Solute carrier family 22 member 1 | | Authors: | Zeng, Y.C, Sobti, M, Stewart, A.G. | | Deposit date: | 2023-04-04 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis of promiscuous substrate transport by Organic Cation Transporter 1.
Nat Commun, 14, 2023
|
|
6KD0
 
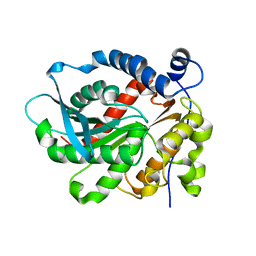 | | Crystal Structure of Vibralactone Cyclase | | Descriptor: | Vibralactone Cyclase | | Authors: | Zeng, Y, Feng, K.N. | | Deposit date: | 2019-06-30 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Hydrolase-Catalyzed Cyclization Forms the Fused Bicyclic beta-Lactone in Vibralactone.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6IYX
 
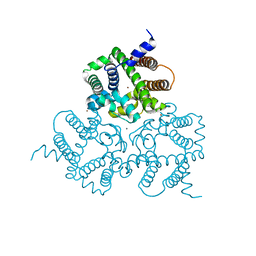 | | Crystal Structure Analysis of a Eukaryotic Membrane Protein | | Descriptor: | BROMIDE ION, CALCIUM ION, Trimeric intracellular cation channel type A | | Authors: | Zeng, Y, Wang, X.H, Gao, F, Su, M, Chen, Y.H. | | Deposit date: | 2018-12-17 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IZ3
 
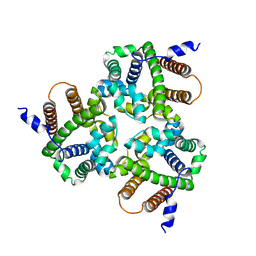 | |
3WRE
 
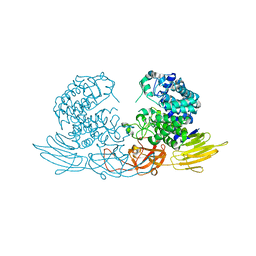 | | The crystal structure of native HypBA1 from Bifidobacterium longum JCM 1217 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
3WRG
 
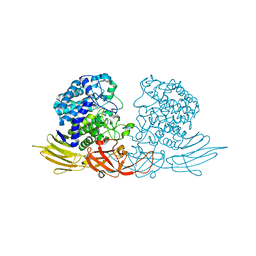 | | The complex structure of HypBA1 with L-arabinose | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase, ZINC ION, beta-L-arabinofuranose | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
3WRF
 
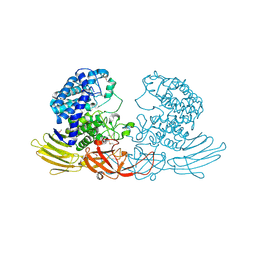 | | The crystal structure of native HypBA1 from Bifidobacterium longum JCM 1217 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
5D0O
 
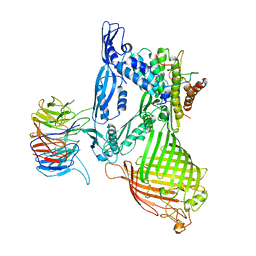 | | BamABCDE complex, outer membrane beta barrel assembly machinery entire complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Gu, Y, Paterson, N, Zeng, Y, Dong, H, Wang, W, Dong, C. | | Deposit date: | 2015-08-03 | | Release date: | 2016-03-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of outer membrane protein insertion by the BAM complex.
Nature, 531, 2016
|
|
5D0Q
 
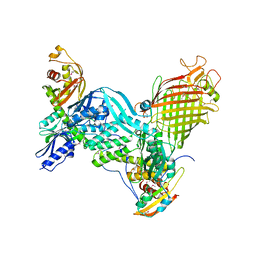 | | BamACDE complex, outer membrane beta-barrel assembly machinery (BAM) complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamC, Outer membrane protein assembly factor BamD, ... | | Authors: | Gu, Y, Paterson, N, Zeng, Y, Dong, H, Wang, W, Dong, C. | | Deposit date: | 2015-08-03 | | Release date: | 2016-03-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of outer membrane protein insertion by the BAM complex.
Nature, 531, 2016
|
|
3JD6
 
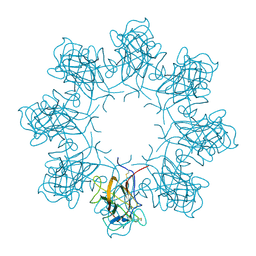 | | Double octamer structure of retinoschisin, a cell-cell adhesion protein of the retina | | Descriptor: | Retinoschisin | | Authors: | Tolun, G, Vijayasarathy, C, Huang, R, Zeng, Y, Li, Y, Steven, A.C, Sieving, P.A, Heymann, J.B. | | Deposit date: | 2016-04-12 | | Release date: | 2016-05-11 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Paired octamer rings of retinoschisin suggest a junctional model for cell-cell adhesion in the retina.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8U1H
 
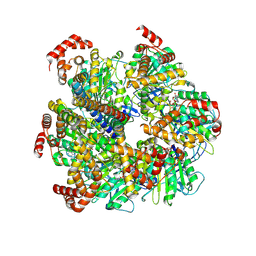 | | Axle-less Bacillus sp. PS3 F1 ATPase mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Furlong, E.J, Zeng, Y.C, Brown, S.H.J, Sobti, M, Stewart, A.G. | | Deposit date: | 2023-09-01 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The molecular structure of an axle-less F1 ATPase
To Be Published
|
|
5WUF
 
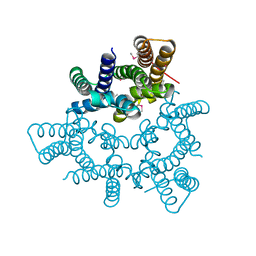 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | CADMIUM ION, Putative membrane protein | | Authors: | Mao, Y, Gao, F, Su, M, Wang, X.H, Zeng, Y, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
6IZF
 
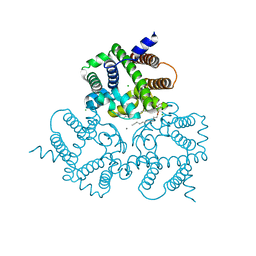 | | Structural basis for activity of TRIC counter-ion channels in calcium release | | Descriptor: | (2R)-1-(dodecanoyloxy)-3-hydroxypropan-2-yl (5E,8E,11E)-tetradeca-5,8,11-trienoate, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, X.H, Zeng, Y, Su, M, Hendrickson, W.A, Chen, Y.H. | | Deposit date: | 2018-12-19 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IYZ
 
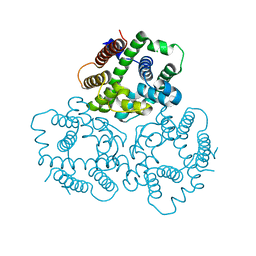 | | Structural basis for activity of TRIC counter-ion channels in calcium release | | Descriptor: | CHLORIDE ION, Trimeric intracellular cation channel type A | | Authors: | Wang, X.H, Zeng, Y, Qin, L, Gao, F, Su, M, Chen, Y.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
