5VGJ
 
 | | Crystal Structure of the Human Fab VRC38.01, an HIV-1 V1V2-Directed Neutralizing Antibody Isolated from Donor N90, bound to a scaffolded WITO V1V2 domain | | 分子名称: | 1FD6-V1V2-WITO, 2-acetamido-2-deoxy-beta-D-glucopyranose, VRC38.01 Fab Heavy Chain, ... | | 著者 | Gorman, J, Li, J, Kwong, P.D. | | 登録日 | 2017-04-11 | | 公開日 | 2017-05-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.456 Å) | | 主引用文献 | Virus-like Particles Identify an HIV V1V2 Apex-Binding Neutralizing Antibody that Lacks a Protruding Loop.
Immunity, 46, 2017
|
|
5VQF
 
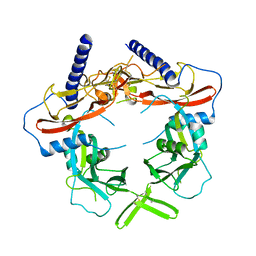 | | Crystal Structure of pro-TGF-beta 1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Transforming growth factor beta-1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Zhao, B, Xu, S, Dong, X, Lu, C, Springer, T.A. | | 登録日 | 2017-05-08 | | 公開日 | 2017-11-15 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Prodomain-growth factor swapping in the structure of pro-TGF-beta 1.
J. Biol. Chem., 293, 2018
|
|
1BIO
 
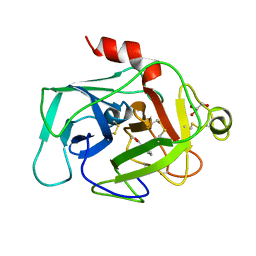 | | HUMAN COMPLEMENT FACTOR D IN COMPLEX WITH ISATOIC ANHYDRIDE INHIBITOR | | 分子名称: | COMPLEMENT FACTOR D, GLYCEROL, ISATOIC ANHYDRIDE | | 著者 | Jing, H, Babu, Y.S, Moore, D, Kilpatrick, J.M, Liu, X.-Y, Volanakis, J.E, Narayana, S.V.L. | | 登録日 | 1998-06-18 | | 公開日 | 1999-06-22 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structures of native and complexed complement factor D: implications of the atypical His57 conformation and self-inhibitory loop in the regulation of specific serine protease activity.
J.Mol.Biol., 282, 1998
|
|
4N7R
 
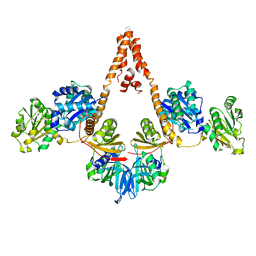 | | Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its binding protein | | 分子名称: | Genomic DNA, chromosome 3, P1 clone: MXL8, ... | | 著者 | Zhao, A, Fang, Y, Lin, Y, Gong, W, Liu, L. | | 登録日 | 2013-10-16 | | 公開日 | 2014-05-14 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.802 Å) | | 主引用文献 | Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its stimulator protein
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8HLL
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 1) | | 分子名称: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | 著者 | Wei, H, Guo, M, Wang, H, Chen, Y. | | 登録日 | 2022-11-30 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8HLN
 
 | | Crystal structure of p53/BCL2 fusion complex(complex3) | | 分子名称: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | 著者 | Guo, M, Wei, H, Wang, H, Chen, Y. | | 登録日 | 2022-11-30 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.354 Å) | | 主引用文献 | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8HLM
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 2) | | 分子名称: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | 著者 | Guo, M, Wang, H, Wei, H, Chen, Y. | | 登録日 | 2022-11-30 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.522 Å) | | 主引用文献 | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
4Q6I
 
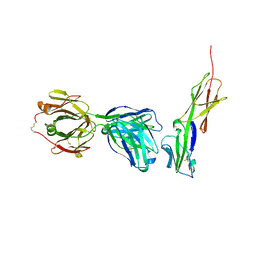 | | Crystal structure of murine 2D5 Fab, a potent anti-CD4 HIV-1-neutralizing antibody in complex with CD4 | | 分子名称: | Heavy chain of murine 2D5 Fab, Light chain of murine 2D5 Fab, T-cell surface glycoprotein CD4 | | 著者 | Boyington, J.C, Nabel, G.J, Mascola, J.R. | | 登録日 | 2014-04-22 | | 公開日 | 2014-07-23 | | 実験手法 | X-RAY DIFFRACTION (3.65 Å) | | 主引用文献 | Neutralizing antibodies to HIV-1 envelope protect more effectively in vivo than those to the CD4 receptor.
Sci Transl Med, 6, 2014
|
|
1DST
 
 | |
4P08
 
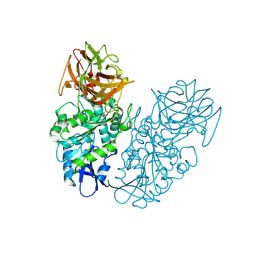 | | Engineered thermostable dimeric cocaine esterase | | 分子名称: | Cocaine esterase | | 著者 | Rodgers, D.W, Chow, K.-M, Fang, L, Zhan, C.-G. | | 登録日 | 2014-02-20 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.341 Å) | | 主引用文献 | Rational design, preparation, and characterization of a therapeutic enzyme mutant with improved stability and function for cocaine detoxification.
Acs Chem.Biol., 9, 2014
|
|
6L6L
 
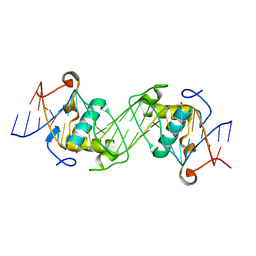 | | Structural basis of NR4A2 homodimers binding to selective Nur-responsive elements | | 分子名称: | DNA (5'-D(*AP*AP*AP*GP*GP*TP*CP*AP*AP*AP*CP*TP*GP*TP*GP*AP*CP*CP*TP*AP*T)-3'), DNA (5'-D(P*TP*AP*TP*AP*GP*GP*TP*CP*AP*CP*AP*GP*TP*TP*TP*GP*AP*CP*CP*TP*T)-3'), Nuclear receptor related 1, ... | | 著者 | Jiang, L, Chen, Y. | | 登録日 | 2019-10-29 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.781 Å) | | 主引用文献 | Structural basis of binding of homodimers of the nuclear receptor NR4A2 to selective Nur-responsive DNA elements.
J.Biol.Chem., 294, 2019
|
|
6L6Q
 
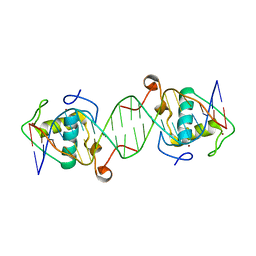 | |
6LBI
 
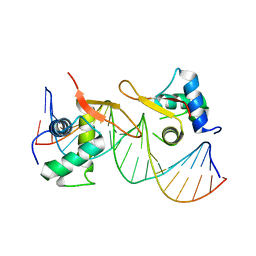 | |
6UG1
 
 | |
6L5M
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 in complex with AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, Nucleolar RNA helicase 2 | | 著者 | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | 登録日 | 2019-10-24 | | 公開日 | 2020-06-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5L
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at apo state | | 分子名称: | MAGNESIUM ION, Nucleolar RNA helicase 2 | | 著者 | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | 登録日 | 2019-10-24 | | 公開日 | 2020-06-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5N
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-unwound state | | 分子名称: | MAGNESIUM ION, Nucleolar RNA helicase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | 登録日 | 2019-10-24 | | 公開日 | 2020-06-17 | | 最終更新日 | 2020-08-12 | | 実験手法 | X-RAY DIFFRACTION (2.242 Å) | | 主引用文献 | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5O
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-hydrolysis state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | 登録日 | 2019-10-24 | | 公開日 | 2020-06-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6LBM
 
 | |
7E3O
 
 | |
7V7L
 
 | | Crystal Structure of the Heterodimeric HIF-3a:ARNT Complex | | 分子名称: | Aryl hydrocarbon receptor nuclear translocator, Hypoxia-inducible factor 3-alpha | | 著者 | Diao, X, Ren, X, Li, F.W, Sun, X, Wu, D. | | 登録日 | 2021-08-21 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Identification of oleoylethanolamide as an endogenous ligand for HIF-3 alpha.
Nat Commun, 13, 2022
|
|
7DTZ
 
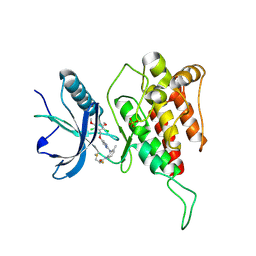 | | FGFR4 complex with a covalent inhibitor | | 分子名称: | Fibroblast growth factor receptor 4, N-[2-[[5-[[2,6-bis(chloranyl)-3,5-dimethoxy-phenyl]methoxy]pyrimidin-2-yl]amino]-3-methyl-phenyl]-2-fluoranyl-prop-2-enamide, SULFATE ION | | 著者 | Chen, X.J, Dai, S.Y, Chen, Y.H. | | 登録日 | 2021-01-07 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Investigation of Covalent Warheads in the Design of 2-Aminopyrimidine-based FGFR4 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7V7W
 
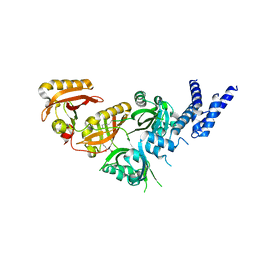 | | Crystal Structure of the Heterodimeric HIF-3a:ARNT Complex with oleoylethanolamide (OEA) | | 分子名称: | (Z)-N-(2-hydroxyethyl)octadec-9-enamide, Aryl hydrocarbon receptor nuclear translocator, Hypoxia-inducible factor 3-alpha | | 著者 | Diao, X, Ren, X, Li, F.W, Zhang, M, Sun, X, Wu, D. | | 登録日 | 2021-08-21 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.507 Å) | | 主引用文献 | Identification of oleoylethanolamide as an endogenous ligand for HIF-3 alpha.
Nat Commun, 13, 2022
|
|
7TCQ
 
 | |
7F7I
 
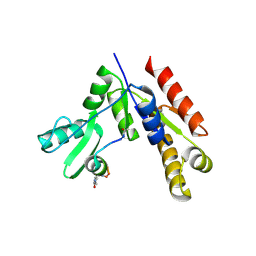 | | Stapled Peptide Inhibitor in complex with PSD95 GK domain | | 分子名称: | ACE-ARG-ILE-ARG-ARG-ASP-GLU-TYR-LEU-LYZ-ALA-ILE-GLN-NH2, Disks large homolog 4 | | 著者 | Shang, Y, Huang, X, Li, X, Zhang, M. | | 登録日 | 2021-06-29 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.595 Å) | | 主引用文献 | Entropy of stapled peptide inhibitors in free state is the major contributor to the improvement of binding affinity with the GK domain.
Rsc Chem Biol, 2, 2021
|
|
