7W0G
 
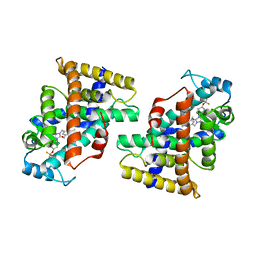 | | Human PPAR delta ligand binding domain in complex with a synthetic agonist H11 | | 分子名称: | 2-[2,6-dimethyl-4-[[5-oxidanylidene-4-[4-(trifluoromethyloxy)phenyl]-1,2,4-triazol-1-yl]methyl]phenoxy]-2-methyl-propanoic acid, Peroxisome proliferator-activated receptor delta | | 著者 | Dai, L, Sun, H.B, Yuan, H.L, Feng, Z.Q. | | 登録日 | 2021-11-18 | | 公開日 | 2022-02-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.443 Å) | | 主引用文献 | Design, Synthesis, and Biological Evaluation of Triazolone Derivatives as Potent PPAR alpha / delta Dual Agonists for the Treatment of Nonalcoholic Steatohepatitis.
J.Med.Chem., 65, 2022
|
|
1XX2
 
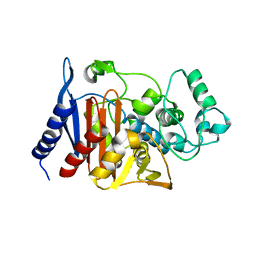 | | Refinement of P99 beta-lactamase from Enterobacter cloacae | | 分子名称: | Beta-lactamase | | 著者 | Knox, J.R, Sun, T. | | 登録日 | 2004-11-03 | | 公開日 | 2004-11-23 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Crystallographic structure of a phosphonate derivative of the Enterobacter cloacae P99 cephalosporinase: mechanistic interpretation of a beta-lactamase transition-state analog.
Biochemistry, 33, 1994
|
|
2HHN
 
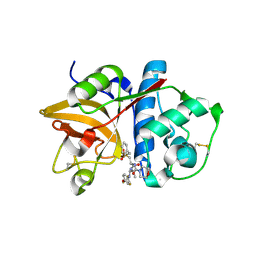 | | Cathepsin S in complex with non covalent arylaminoethyl amide. | | 分子名称: | Cathepsin S, N-[(1R)-1-[(BENZYLSULFONYL)METHYL]-2-{[(1S)-1-METHYL-2-{[4-(TRIFLUOROMETHOXY)PHENYL]AMINO}ETHYL]AMINO}-2-OXOETHYL]MORPHOLINE-4-CARBOXAMIDE, SULFATE ION | | 著者 | Spraggon, G, Hornsby, M, Lesley, S.A, Tully, D.C, Harris, J.L, Karenewsky, D.S, Kulathila, R, Clark, K. | | 登録日 | 2006-06-28 | | 公開日 | 2007-05-08 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Synthesis and SAR of arylaminoethyl amides as noncovalent inhibitors of cathepsin S: P3 cyclic ethers
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2ICZ
 
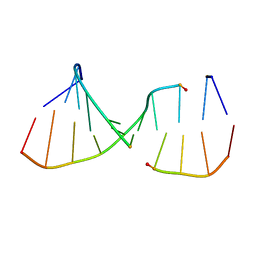 | | NMR Structures of the Expanded DNA 10bp xTGxTAxCxGCxAxGT:xACTxGCGxTAxCA | | 分子名称: | 5'-D(*(XAE)P*CP*TP*(XGA)P*CP*GP*(XTY)P*AP*(XCS)P*A)-3', 5'-D(*(XTY)P*GP*(XTY)P*AP*(XCS)P*(XGA)P*CP*(XAE)P*(XGA)P*T)-3' | | 著者 | Lynch, S.R. | | 登録日 | 2006-09-13 | | 公開日 | 2006-11-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Toward a Designed, Functioning Genetic System with Expanded-Size Base Pairs: Solution Structure of the Eight-Base xDNA Double Helix.
J.Am.Chem.Soc., 128, 2006
|
|
2JSW
 
 | | Solution Structure of the R13 Domain of Talin | | 分子名称: | Talin-1 | | 著者 | Goult, B.T, Gingras, A.R, Bate, N, Critchley, D.R.C, Barsukov, I.L. | | 登録日 | 2007-07-17 | | 公開日 | 2008-01-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structure of the C-terminal actin-binding domain of talin.
Embo J., 27, 2007
|
|
7DNC
 
 | | Crystal structure of EV71 3C proteinase in complex with a novel inhibitor | | 分子名称: | 3C protease, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | 著者 | Xie, H, Su, H.X, Li, M.J, Xu, Y.C. | | 登録日 | 2020-12-09 | | 公開日 | 2021-05-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.17 Å) | | 主引用文献 | Design, Synthesis, and Biological Evaluation of Peptidomimetic Aldehydes as Broad-Spectrum Inhibitors against Enterovirus and SARS-CoV-2.
J.Med.Chem., 65, 2022
|
|
5XEG
 
 | | The structure of OsALKBH1 | | 分子名称: | 2-OXOGLUTARIC ACID, MANGANESE (II) ION, Oxidoreductase, ... | | 著者 | Wang, C, Guo, Y, Zeng, Z. | | 登録日 | 2017-04-05 | | 公開日 | 2018-06-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Identification and analysis of adenine N6-methylation sites in the rice genome.
Nat Plants, 4, 2018
|
|
7CBQ
 
 | | Crystal structure of PDE4D catalytic domain in complex with Apremilast | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, N-{2-[(1S)-1-(3-ethoxy-4-methoxyphenyl)-2-(methylsulfonyl)ethyl]-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl}acetamide, ... | | 著者 | Zhang, X.L, Xu, Y.C. | | 登録日 | 2020-06-13 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Design, synthesis, and biological evaluation of tetrahydroisoquinolines derivatives as novel, selective PDE4 inhibitors for antipsoriasis treatment.
Eur.J.Med.Chem., 211, 2021
|
|
8HFQ
 
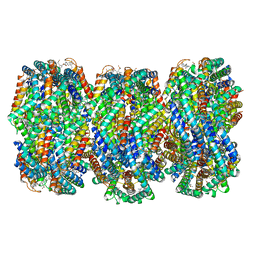 | | Cryo-EM structure of CpcL-PBS from cyanobacterium Synechocystis sp. PCC 6803 | | 分子名称: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, Ferredoxin--NADP reductase, ... | | 著者 | Zheng, L, Zhang, Z, Wang, H, Zheng, Z, Gao, N, Zhao, J. | | 登録日 | 2022-11-11 | | 公開日 | 2023-11-08 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | Cryo-EM and femtosecond spectroscopic studies provide mechanistic insight into the energy transfer in CpcL-phycobilisomes.
Nat Commun, 14, 2023
|
|
7CBJ
 
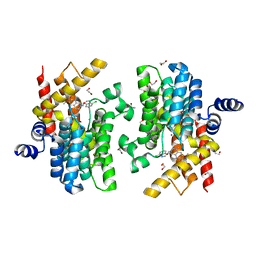 | | Crystal structure of PDE4D catalytic domain in complex with compound 36 | | 分子名称: | (1S)-1-[(7-chloranyl-1H-indol-3-yl)methyl]-6,7-dimethoxy-3,4-dihydro-1H-isoquinoline-2-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | 著者 | Zhang, X.L, Xu, Y.C. | | 登録日 | 2020-06-12 | | 公開日 | 2021-06-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Design, synthesis, and biological evaluation of tetrahydroisoquinolines derivatives as novel, selective PDE4 inhibitors for antipsoriasis treatment.
Eur.J.Med.Chem., 211, 2021
|
|
8I8Y
 
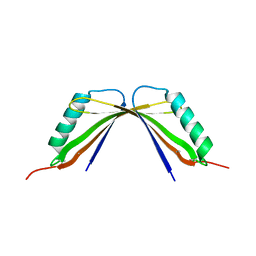 | | A mutant of the C-terminal complex of proteins 4.1G and NuMA | | 分子名称: | Engineered protein | | 著者 | Hu, X. | | 登録日 | 2023-02-06 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Combined prediction and design reveals the target recognition mechanism of an intrinsically disordered protein interaction domain.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7DMF
 
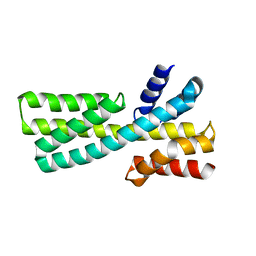 | |
7E9G
 
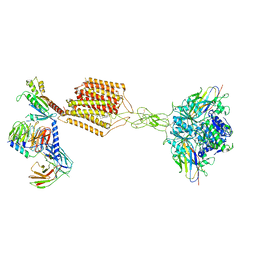 | | Cryo-EM structure of Gi-bound metabotropic glutamate receptor mGlu2 | | 分子名称: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 1-butyl-3-chloranyl-4-(4-phenylpiperidin-1-yl)pyridin-2-one, DN13, ... | | 著者 | Lin, S, Han, S, Zhao, Q, Wu, B. | | 登録日 | 2021-03-04 | | 公開日 | 2021-06-23 | | 最終更新日 | 2021-08-04 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structures of G i -bound metabotropic glutamate receptors mGlu2 and mGlu4.
Nature, 594, 2021
|
|
7E9H
 
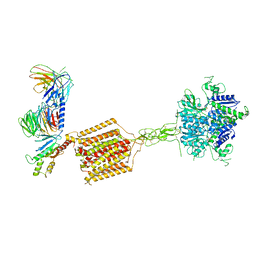 | | Cryo-EM structure of Gi-bound metabotropic glutamate receptor mGlu4 | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-3, ... | | 著者 | Lin, S, Han, S, Zhao, Q, Wu, B. | | 登録日 | 2021-03-04 | | 公開日 | 2021-06-23 | | 最終更新日 | 2021-07-07 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structures of G i -bound metabotropic glutamate receptors mGlu2 and mGlu4.
Nature, 594, 2021
|
|
4K6J
 
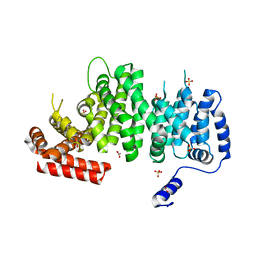 | | Human cohesin inhibitor WapL | | 分子名称: | ACETATE ION, SULFATE ION, Wings apart-like protein homolog | | 著者 | Tomchick, D.R, Yu, H, Ouyang, Z. | | 登録日 | 2013-04-16 | | 公開日 | 2013-06-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6205 Å) | | 主引用文献 | Structure of the human cohesin inhibitor Wapl.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5YAG
 
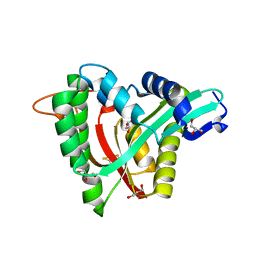 | | Crystal structure of mosquito arylalkylamine N-Acetyltransferase like 5b/spermine N-Acetyltransferase | | 分子名称: | AAEL004827-PA, GLYCEROL | | 著者 | Han, Q, Guan, H, Robinson, H, Li, J. | | 登録日 | 2017-08-31 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Identification of aaNAT5b as a spermine N-acetyltransferase in the mosquito, Aedes aegypti.
PLoS ONE, 13, 2018
|
|
4AFN
 
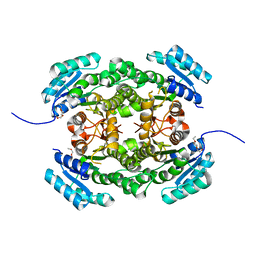 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa at 2.3A resolution | | 分子名称: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, PENTAETHYLENE GLYCOL | | 著者 | Cukier, C.D, Schnell, R, Schneider, G, Lindqvist, Y. | | 登録日 | 2012-01-20 | | 公開日 | 2013-01-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
8I71
 
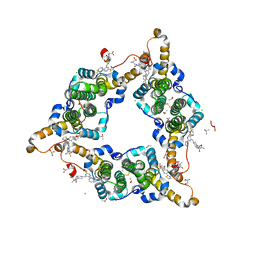 | | Hepatitis B virus core protein Y132A mutant in complex with Linvencorvir (RG7907), a Hepatitis B Virus (HBV) Core Protein Allosteric Modulator (CpAM) | | 分子名称: | 3-[(8~{a}~{S})-7-[[5-ethoxycarbonyl-4-(3-fluoranyl-2-methyl-phenyl)-2-(1,3-thiazol-2-yl)-1,4-dihydropyrimidin-6-yl]methyl]-3-oxidanylidene-5,6,8,8~{a}-tetrahydro-1~{H}-imidazo[1,5-a]pyrazin-2-yl]-2,2-dimethyl-propanoic acid, CHLORIDE ION, Capsid protein, ... | | 著者 | Zhou, Z, Xu, Z.H. | | 登録日 | 2023-01-30 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-04-05 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Discovery of Linvencorvir (RG7907), a Hepatitis B Virus Core Protein Allosteric Modulator, for the Treatment of Chronic HBV Infection.
J.Med.Chem., 66, 2023
|
|
8HTV
 
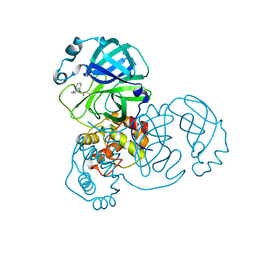 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3a | | 分子名称: | 1-(5,6-dihydrobenzo[b][1]benzazepin-11-yl)-2-sulfanyl-ethanone, 3C-like proteinase | | 著者 | Su, H.X, Nie, T.Q, Li, M.J, Xu, Y.C. | | 登録日 | 2022-12-21 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Discovery and Mechanism Study of SARS-CoV-2 3C-like Protease Inhibitors with a New Reactive Group.
J.Med.Chem., 66, 2023
|
|
4AG3
 
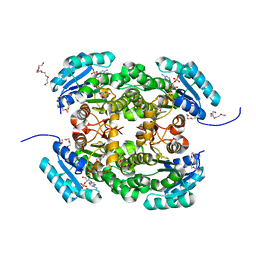 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with NADPH at 1.8A resolution | | 分子名称: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PENTAETHYLENE GLYCOL | | 著者 | Cukier, C.D, Schnell, R, Schneider, G, Lindqvist, Y. | | 登録日 | 2012-01-24 | | 公開日 | 2013-02-06 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4LWF
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ3 | | 分子名称: | 4-(5-amino-1,2-oxazol-3-yl)-6-(propan-2-yl)benzene-1,3-diol, Heat shock protein HSP 90-alpha | | 著者 | Li, J, Shi, F, Xiong, B, He, J. | | 登録日 | 2013-07-27 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
4LWH
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ5 | | 分子名称: | Heat shock protein HSP 90-alpha, N-{3-[2,4-dihydroxy-5-(propan-2-yl)phenyl]-4-[4-(morpholin-4-ylmethyl)phenyl]-1,2-oxazol-5-yl}cyclopropanecarboxamide | | 著者 | Li, J, Shi, F, Xiong, B, He, J. | | 登録日 | 2013-07-27 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
4LWG
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ4 | | 分子名称: | 1-(5-chloro-2,4-dihydroxyphenyl)-2-(4-methoxyphenyl)ethanone, Heat shock protein HSP 90-alpha | | 著者 | Li, J, Shi, F, Xiong, B, He, J. | | 登録日 | 2013-07-27 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.599 Å) | | 主引用文献 | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
6AKY
 
 | | The Crystal structure of Human Chemokine Receptor CCR5 in complex with compound 34 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4,4-difluoro-N-[(1S)-3-{(3-exo)-3-[3-methyl-5-(propan-2-yl)-4H-1,2,4-triazol-4-yl]-8-azabicyclo[3.2.1]octan-8-yl}-1-(thiophen-3-yl)propyl]cyclohexane-1-carboxamide, C-C chemokine receptor type 5,Rubredoxin,C-C chemokine receptor type 5, ... | | 著者 | Zhu, Y, Zhao, Q, Wu, B. | | 登録日 | 2018-09-04 | | 公開日 | 2018-10-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure-Based Design of 1-Heteroaryl-1,3-propanediamine Derivatives as a Novel Series of CC-Chemokine Receptor 5 Antagonists.
J. Med. Chem., 61, 2018
|
|
6AKS
 
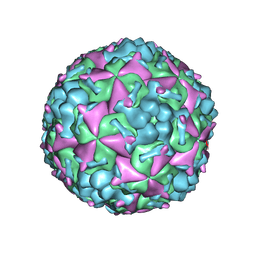 | | Cryo-EM structure of CVA10 mature virus | | 分子名称: | SPHINGOSINE, VP1, VP2, ... | | 著者 | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | 登録日 | 2018-09-03 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
