8GK4
 
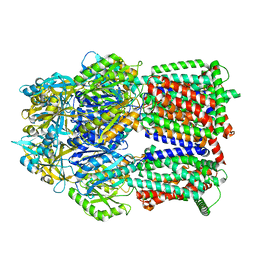 | | Multi-drug efflux pump RE-CmeB bound with Chloramphenicol | | 分子名称: | CHLORAMPHENICOL, Efflux pump membrane transporter | | 著者 | Zhang, Z. | | 登録日 | 2023-03-17 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (3.12 Å) | | 主引用文献 | Cryo-Electron Microscopy Structures of a Campylobacter Multidrug Efflux Pump Reveal a Novel Mechanism of Drug Recognition and Resistance.
Microbiol Spectr, 11, 2023
|
|
8GJK
 
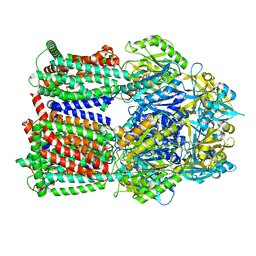 | | Multi-drug efflux pump RE-CmeB bound with ampicillin | | 分子名称: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Efflux pump membrane transporter | | 著者 | Zhang, Z. | | 登録日 | 2023-03-16 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Cryo-Electron Microscopy Structures of a Campylobacter Multidrug Efflux Pump Reveal a Novel Mechanism of Drug Recognition and Resistance.
Microbiol Spectr, 11, 2023
|
|
8GJL
 
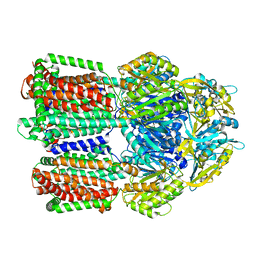 | | multi-drug efflux pump RE-CmeB bound with Ciprofloxacin | | 分子名称: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, Efflux pump membrane transporter | | 著者 | Zhang, Z. | | 登録日 | 2023-03-16 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (3.44 Å) | | 主引用文献 | Cryo-Electron Microscopy Structures of a Campylobacter Multidrug Efflux Pump Reveal a Novel Mechanism of Drug Recognition and Resistance.
Microbiol Spectr, 11, 2023
|
|
8GJJ
 
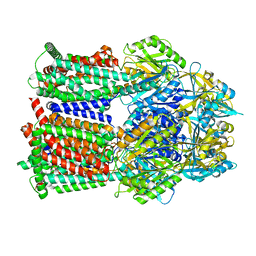 | | Multi-drug efflux pump RE-CmeB Apo form | | 分子名称: | Efflux pump membrane transporter | | 著者 | Zhang, Z. | | 登録日 | 2023-03-15 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Cryo-Electron Microscopy Structures of a Campylobacter Multidrug Efflux Pump Reveal a Novel Mechanism of Drug Recognition and Resistance.
Microbiol Spectr, 11, 2023
|
|
8GK0
 
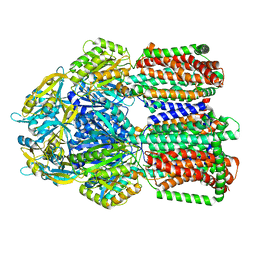 | | Multi-drug efflux pump RE-CmeB bound with Erythromycin | | 分子名称: | ERYTHROMYCIN A, Efflux pump membrane transporter | | 著者 | Zhang, Z. | | 登録日 | 2023-03-16 | | 公開日 | 2023-06-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (3.44 Å) | | 主引用文献 | Cryo-Electron Microscopy Structures of a Campylobacter Multidrug Efflux Pump Reveal a Novel Mechanism of Drug Recognition and Resistance.
Microbiol Spectr, 11, 2023
|
|
7JUN
 
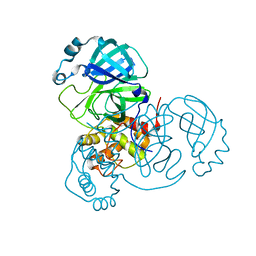 | |
7CZ6
 
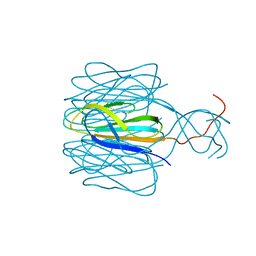 | |
7D0L
 
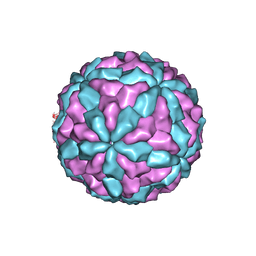 | | The major capsid of Omono River virus (strain:LZ), protrusion-free status. | | 分子名称: | Capsid protein | | 著者 | Shao, Q, Jia, X, Gao, Y, Liu, Z. | | 登録日 | 2020-09-11 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | Cryo-EM reveals a previously unrecognized structural protein of a dsRNA virus implicated in its extracellular transmission.
Plos Pathog., 17, 2021
|
|
7D0K
 
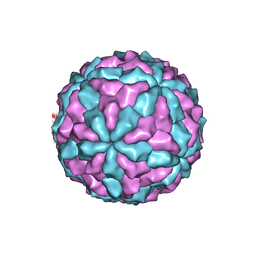 | |
7KJS
 
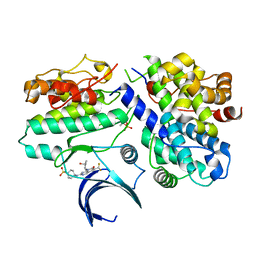 | | Crystal structure of CDK2/cyclin E in complex with PF-06873600 | | 分子名称: | 6-(difluoromethyl)-8-[(1R,2R)-2-hydroxy-2-methylcyclopentyl]-2-{[1-(methylsulfonyl)piperidin-4-yl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | 著者 | McTigue, M.A, He, Y, Ferre, R.A. | | 登録日 | 2020-10-26 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.187 Å) | | 主引用文献 | Discovery of PF-06873600, a CDK2/4/6 Inhibitor for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
7XMS
 
 | | CryoEM structure of somatostatin receptor 4 (SSTR4) in complex with Gi1 and its endogeneous ligand SST-14 | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | 登録日 | 2022-04-26 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XMT
 
 | | CryoEM structure of somatostatin receptor 4 (SSTR4) with Gi1 and J-2156 | | 分子名称: | (2~{S})-2-[[(2~{S})-4-azanyl-2-[(4-methylnaphthalen-1-yl)sulfonylamino]butanoyl]amino]-3-phenyl-propanimidic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | 登録日 | 2022-04-26 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XMR
 
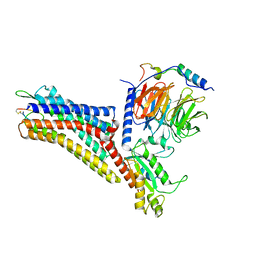 | | CryoEM structure of the somatostatin receptor 2 (SSTR2) in complex with Gi1 and its endogeneous peptide ligand SST-14 | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | 登録日 | 2022-04-26 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
2GA2
 
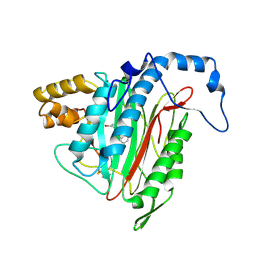 | | h-MetAP2 complexed with A193400 | | 分子名称: | 5-BROMO-2-{[(4-CHLOROPHENYL)SULFONYL]AMINO}BENZOIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | 著者 | Park, C. | | 登録日 | 2006-03-07 | | 公開日 | 2007-03-13 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Development of sulfonamide compounds as potent methionine aminopeptidase type II inhibitors with antiproliferative properties.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
7BVH
 
 | | Crystal structure of arabinosyltransferase EmbC2-AcpM2 complex from Mycobacterium smegmatis complexed with di-arabinose | | 分子名称: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, Meromycolate extension acyl carrier protein, ... | | 著者 | Zhao, Y, Zhang, L, Wu, L.J, Wang, Q, Li, J, Besra, G.S, Rao, Z.H. | | 登録日 | 2020-04-10 | | 公開日 | 2020-04-29 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVF
 
 | | Cryo-EM structure of Mycobacterium tuberculosis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | 分子名称: | CALCIUM ION, CARDIOLIPIN, Ethambutol, ... | | 著者 | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | 登録日 | 2020-04-10 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.97 Å) | | 主引用文献 | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVC
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | 分子名称: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | 著者 | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | 登録日 | 2020-04-10 | | 公開日 | 2020-04-29 | | 最終更新日 | 2020-07-01 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVE
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbC2-AcpM2 in complex with ethambutol | | 分子名称: | CALCIUM ION, Ethambutol, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, ... | | 著者 | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | 登録日 | 2020-04-10 | | 公開日 | 2020-04-29 | | 最終更新日 | 2020-07-01 | | 実験手法 | ELECTRON MICROSCOPY (2.81 Å) | | 主引用文献 | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVG
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with di-arabinose. | | 分子名称: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | 著者 | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | 登録日 | 2020-04-10 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7CJF
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody heavy chain, ... | | 著者 | Guo, Y, Li, X, Zhang, G, Fu, D, Schweizer, L, Zhang, H, Rao, Z. | | 登録日 | 2020-07-10 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.108 Å) | | 主引用文献 | A SARS-CoV-2 neutralizing antibody with extensive Spike binding coverage and modified for optimal therapeutic outcomes.
Nat Commun, 12, 2021
|
|
4N0N
 
 | | Crystal structure of Arterivirus nonstructural protein 10 (helicase) | | 分子名称: | MAGNESIUM ION, Replicase polyprotein 1ab, SULFATE ION, ... | | 著者 | Deng, Z, Chen, Z. | | 登録日 | 2013-10-02 | | 公開日 | 2014-01-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase.
Nucleic Acids Res., 42, 2014
|
|
4N0O
 
 | |
7V2A
 
 | | SARS-CoV-2 Spike trimer in complex with XG014 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, The heavy chain of XG014, ... | | 著者 | Wang, K, Wang, X, Pan, L. | | 登録日 | 2021-08-07 | | 公開日 | 2021-10-20 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
7E8M
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1F11 with mutated RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1F11 heavy chain, ... | | 著者 | Wang, X.Q, Zhang, L.Q, Ge, J.W, Wang, R.K, Lan, J. | | 登録日 | 2021-03-02 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species.
Immunity, 54, 2021
|
|
4R91
 
 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-4-(((1S,3R)-3-(cyclopentylamino)cyclohexyl)methyl)-1-methyl-5-oxoimidazolidin-2-iminium | | 分子名称: | (2E,5R)-5-(2-cyclohexylethyl)-5-{[(1S,3R)-3-(cyclopentylamino)cyclohexyl]methyl}-2-imino-3-methylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Orth, P, Caldwell, J.P, Strickland, C. | | 登録日 | 2014-09-03 | | 公開日 | 2014-11-05 | | 最終更新日 | 2014-12-17 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
