5KAF
 
 | | RT XFEL structure of Photosystem II in the dark state at 3.0 A resolution | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | 登録日 | 2016-06-01 | | 公開日 | 2016-11-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.00001 Å) | | 主引用文献 | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5KAI
 
 | | NH3-bound RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 2.8 A resolution | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | 登録日 | 2016-06-01 | | 公開日 | 2016-11-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.80000925 Å) | | 主引用文献 | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
6W05
 
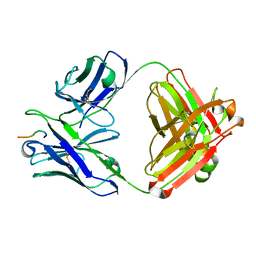 | |
6W00
 
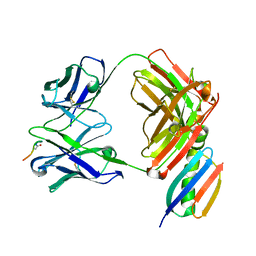 | | Crystal structure of Fab239 in complex with NPNA2 peptide from circumsporozoite protein | | 分子名称: | Fab239 heavy chain, Fab239 light chain, Immunoglobulin G-binding protein G, ... | | 著者 | Pholcharee, T, Oyen, D, Wilson, I.A. | | 登録日 | 2020-02-28 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.853 Å) | | 主引用文献 | Structural and biophysical correlation of anti-NANP antibodies with in vivo protection against P. falciparum.
Nat Commun, 12, 2021
|
|
6WG1
 
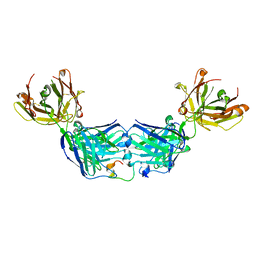 | |
4URV
 
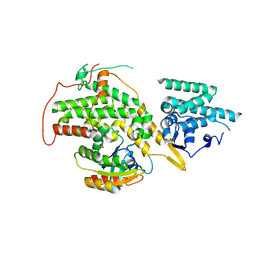 | | The crystal structure of H-Ras and SOS in complex with ligands | | 分子名称: | 4-(4-BROMOPHENYL)PIPERIDIN-4-OL, FORMIC ACID, GTPASE HRAS, ... | | 著者 | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | 登録日 | 2014-07-02 | | 公開日 | 2015-03-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
4US1
 
 | | The crystal structure of H-Ras and SOS in complex with ligands | | 分子名称: | (3S)-3-[3-(aminomethyl)phenyl]-1-ethylpyrrolidine-2,5-dione, GTPASE HRAS, SON OF SEVENLESS HOMOLOG 1 | | 著者 | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | 登録日 | 2014-07-02 | | 公開日 | 2015-03-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
4URW
 
 | | The crystal structure of H-Ras and SOS in complex with ligands | | 分子名称: | 2-(2,6-DIMETHYLPHENYL)-4-(METHYLSULFANYL)-6-(PIPERAZIN-1-YL)-1,3,5-TRIAZINE, GTPASE HRAS, SON OF SEVENLESS HOMOLOG 1 | | 著者 | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | 登録日 | 2014-07-02 | | 公開日 | 2015-03-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
4URY
 
 | | The crystal structure of H-Ras and SOS in complex with ligands | | 分子名称: | GTPASE HRAS, N-[(4-aminophenyl)sulfonyl]cyclopropanecarboxamide, SON OF SEVENLESS HOMOLOG 1 | | 著者 | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | 登録日 | 2014-07-02 | | 公開日 | 2015-03-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
122D
 
 | |
123D
 
 | |
5K3Y
 
 | | Crystal structure of AuroraB/INCENP in complex with BI 811283 | | 分子名称: | Aurora kinase B-A, Inner centromere protein A, N-methyl-N-(1-methylpiperidin-4-yl)-4-{[4-({(1R,2S)-2-[(propan-2-yl)carbamoyl]cyclopentyl}amino)-5-(trifluoromethyl)pyrimidin-2-yl]amino}benzamide | | 著者 | Bader, G, Zahn, S.K, Zoephel, A. | | 登録日 | 2016-05-20 | | 公開日 | 2016-08-17 | | 最終更新日 | 2022-12-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Pharmacological Profile of BI 847325, an Orally Bioavailable, ATP-Competitive Inhibitor of MEK and Aurora Kinases.
Mol.Cancer Ther., 15, 2016
|
|
4IEH
 
 | |
5DE1
 
 | | Crystal structure of human IDH1 in complex with GSK321A | | 分子名称: | (7R)-1-(4-fluorobenzyl)-N-{3-[(1S)-1-hydroxyethyl]phenyl}-7-methyl-5-(1H-pyrrol-2-ylcarbonyl)-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridine-3-carboxamide, Isocitrate dehydrogenase [NADP] cytoplasmic, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Concha, N.O, Smallwood, A, Qi, H. | | 登録日 | 2015-08-25 | | 公開日 | 2015-10-07 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | New IDH1 mutant inhibitors for treatment of acute myeloid leukemia.
Nat.Chem.Biol., 11, 2015
|
|
4RVW
 
 | |
5KAE
 
 | |
8W0O
 
 | | GDH-105 crystal structure | | 分子名称: | CHLORIDE ION, Dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Peat, T.S, Newman, J. | | 登録日 | 2024-02-13 | | 公開日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Enhancing the Imine Reductase Activity of a Promiscuous Glucose Dehydrogenase for Scalable Manufacturing of a Chiral Neprilysin Inhibitor Precursor
Acs Catalysis, 14, 2024
|
|
8W0N
 
 | | IRED crystal structure | | 分子名称: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, IRED, ... | | 著者 | Peat, T.S, Newman, J. | | 登録日 | 2024-02-13 | | 公開日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | Enhancing the Imine Reductase Activity of a Promiscuous Glucose Dehydrogenase for Scalable Manufacturing of a Chiral Neprilysin Inhibitor Precursor
Acs Catalysis, 14, 2024
|
|
4UW1
 
 | | X-ray crystal structure of human TNKS in complex with a small molecule inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 3-{4-[(dimethylamino)methyl]phenyl}-5-methoxyisoquinolin-1(2H)-one, GLYCEROL, ... | | 著者 | Oliver, A.W, Rajasekaran, M.B, Pearl, L.H. | | 登録日 | 2014-08-08 | | 公開日 | 2015-07-08 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.37 Å) | | 主引用文献 | Design and Discovery of 3-Aryl-5-Substituted-Isoquinolin-1-Ones as Potent and Selective Tankyrase Inhibitors
Medchemcommm, 6, 2015
|
|
6J04
 
 | |
6PLA
 
 | | Adducts formed after 1 month in the reaction of dichlorido(1,3-dimethylbenzimidazol-2-ylidene)(eta6-p-cymene)osmium(II) with HEWL | | 分子名称: | Lysozyme, OSMIUM ION, SODIUM ION, ... | | 著者 | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | 登録日 | 2019-06-30 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Probing the Paradigm of Promiscuity for N-Heterocyclic Carbene Complexes and their Protein Adduct Formation.
Angew.Chem.Int.Ed.Engl., 2021
|
|
6PLB
 
 | | Adducts formed after 1 month in the reaction of dichlorido(1,3-dimethylbenzimida zol-2-ylidene)(eta5-pentamethylcyclopentadienyl)iridium(III) with HEWL | | 分子名称: | 2-(1-chloranyl-2,3,4,5,6-pentamethyl-1$l^{7}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexan-1-yl)-1,3-dimethyl-benzimidazole, Lysozyme, SODIUM ION | | 著者 | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | 登録日 | 2019-06-30 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Probing the Paradigm of Promiscuity for N-Heterocyclic Carbene Complexes and their Protein Adduct Formation.
Angew.Chem.Int.Ed.Engl., 2021
|
|
7B5K
 
 | | E. coli 70S containing suppressor tRNA in the A-site stabilized by a Negamycin analogue and P-site tRNA-nascent chain. | | 分子名称: | 16S rRNA, 2-[[[(3~{R},5~{R})-3-azanyl-6-(3-azanylpropylamino)-5-oxidanyl-hexanoyl]amino]-methyl-amino]ethanoic acid, 23S rRNA, ... | | 著者 | Albers, S, Beckert, B, Matthies, M, Schuster, R, Riedner, M, Seuring, C, Sanyal, S, Torda, A, Wilson, N.D, Ignatova, Z. | | 登録日 | 2020-12-03 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Harnessing evolutionarily flexibility in tRNAs to maximize nonsense suppression activities
Nat Commun, 2021
|
|
7T0O
 
 | |
7T0R
 
 | | Crystal structure of the anti-CD4 adnectin 6940_B01 as a complex with the extracellular domains of CD4 and ibalizumab fAb | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adnectin 6940_B01, Ibalizumab Heavy Chain, ... | | 著者 | Williams, S.P, Concha, N.O, Wensel, D.L, Hong, X. | | 登録日 | 2021-11-30 | | 公開日 | 2022-01-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.65 Å) | | 主引用文献 | Novel Bent Conformation of CD4 Induced by HIV-1 Inhibitor Indirectly Prevents Productive Viral Attachment.
J.Mol.Biol., 434, 2021
|
|
