5KF4
 
 | |
8CTG
 
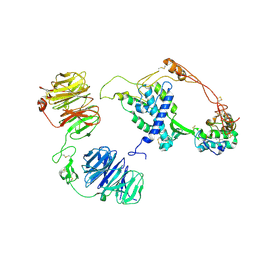 | | Extracellular architecture of an engineered canonical Wnt signaling ternary complex | | Descriptor: | Frizzled-8, Low-density lipoprotein receptor-related protein 6, PALMITOLEIC ACID, ... | | Authors: | Tsutsumi, N, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-14 | | Release date: | 2023-03-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Wnt-Frizzled-LRP6 initiation complex reveals the basis for coreceptor discrimination.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FFE
 
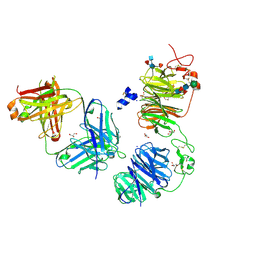 | | Crystal structure of LRP6 E1E2 domains bound to YW210.09 Fab and engineered XWnt8 peptide | | Descriptor: | GLYCEROL, Low-density lipoprotein receptor-related protein 6, SODIUM ION, ... | | Authors: | Jude, K.M, Tsutsumi, N, Waghray, D, Garcia, K.C. | | Deposit date: | 2022-12-08 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure of the Wnt-Frizzled-LRP6 initiation complex reveals the basis for coreceptor discrimination.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HLE
 
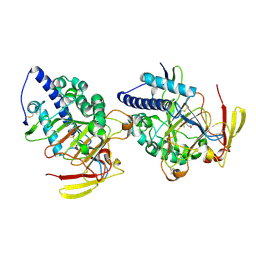 | | Structure of DddY-DMSOP complex | | Descriptor: | 3-[dimethyl(oxidanyl)-$l^{4}-sulfanyl]propanoic acid, DMSP lyase DddY, ZINC ION | | Authors: | Peng, M, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2022-11-30 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | DMSOP-cleaving enzymes are diverse and widely distributed in marine microorganisms.
Nat Microbiol, 8, 2023
|
|
8HLF
 
 | | Crystal structure of DddK-DMSOP complex | | Descriptor: | 3-[dimethyl(oxidanyl)-$l^{4}-sulfanyl]propanoic acid, MANGANESE (II) ION, Novel protein with potential Cupin domain | | Authors: | Peng, M, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2022-11-30 | | Release date: | 2023-10-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | DMSOP-cleaving enzymes are diverse and widely distributed in marine microorganisms.
Nat Microbiol, 8, 2023
|
|
9LTK
 
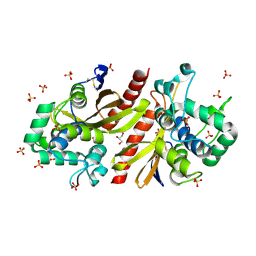 | | Crystal structur of Lpg1618(R215F) from Legionella pneumophila | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Liu, T, Gao, J, Ge, H. | | Deposit date: | 2025-02-06 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and molecular characterization of AmpS, a class D beta-lactamase from Legionella pneumophila.
Int.J.Biol.Macromol., 312, 2025
|
|
6KVF
 
 | | Structure of anti-hCXCR2 abN48 in complex with its CXCR2 epitope | | Descriptor: | Peptide from C-X-C chemokine receptor type 2, heavy chain, light chain | | Authors: | Xiang, J.C, Yan, L, Yang, B, Wilson, I.A. | | Deposit date: | 2019-09-04 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Selection of a picomolar antibody that targets CXCR2-mediated neutrophil activation and alleviates EAE symptoms.
Nat Commun, 12, 2021
|
|
9LJJ
 
 | | cryo-EM structure of a nanobody bound heliorhodopsin | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, ACETATE ION, Heliorhodopsin, ... | | Authors: | He, Y, Xia, R. | | Deposit date: | 2025-01-15 | | Release date: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Cryo-EM structure of a nanobody-bound heliorhodopsin.
Biochem.Biophys.Res.Commun., 750, 2025
|
|
6XKL
 
 | | SARS-CoV-2 HexaPro S One RBD up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wrapp, D, Hsieh, C.-L, Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structure-based design of prefusion-stabilized SARS-CoV-2 spikes.
Science, 369, 2020
|
|
4ZS6
 
 | | Receptor binding domain and Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein, fab Heavy Chain, ... | | Authors: | Yu, X, Wang, X. | | Deposit date: | 2015-05-13 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.166 Å) | | Cite: | Structural basis for the neutralization of MERS-CoV by a human monoclonal antibody MERS-27
Sci Rep, 5, 2015
|
|
6C6X
 
 | | Crystal structure of Middle-East Respiratory Syndrome (MERS) coronavirus neutralizing antibody JC57-14 isolated from a vaccinated rhesus macaque. | | Descriptor: | JC57-14 Heavy chain, JC57-14 Light chain | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2018-01-19 | | Release date: | 2018-02-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Importance of Neutralizing Monoclonal Antibodies Targeting Multiple Antigenic Sites on the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein To Avoid Neutralization Escape.
J. Virol., 92, 2018
|
|
6C6Y
 
 | | Crystal structure of Middle-East Respiratory Syndrome (MERS) coronavirus neutralizing antibody JC57-14 isolated from a vaccinated rhesus macaque in complex with MERS Receptor Binding Domain | | Descriptor: | JC57-14 Heavy chain, JC57-14 Light chain, SULFATE ION, ... | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2018-01-19 | | Release date: | 2018-03-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Importance of Neutralizing Monoclonal Antibodies Targeting Multiple Antigenic Sites on the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein To Avoid Neutralization Escape.
J. Virol., 92, 2018
|
|
7RWE
 
 | | Crystal structure of CDK2 liganded with compound GPHR787 | | Descriptor: | 5-nitro-2-[(3-phenylpropyl)amino]benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-08-19 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
7RXO
 
 | | Crystal structure of CDK2 liganded with compound WN333 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-(methoxycarbonyl)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-08-23 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
7S4T
 
 | | Crystal structure of CDK2 liganded with compound EF2252 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(6-chloro-1H-indol-3-yl)ethyl]amino}-5-nitrobenzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
7S7A
 
 | | Crystal structure of CDK2 liganded with compound EF3019 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(2H-indazol-3-yl)ethyl]amino}-5-(trifluoromethyl)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-15 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
7S85
 
 | | Crystal structure of CDK2 liganded with compound WN316 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-(trifluoromethoxy)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
7KBB
 
 | |
7KBA
 
 | |
7M1C
 
 | |
7M30
 
 | |
7LYV
 
 | |
7M22
 
 | |
7LYW
 
 | |
9JMZ
 
 | | Cryo-EM structure of a human-infecting bovine influenza H5N1 hemagglutinin complexed with avian receptor analog LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Wang, H.C, Han, P, Song, H, Gao, G.F. | | Deposit date: | 2024-09-22 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Receptor binding, structure, and tissue tropism of cattle-infecting H5N1 avian influenza virus hemagglutinin.
Cell, 188, 2025
|
|
