6KPC
 
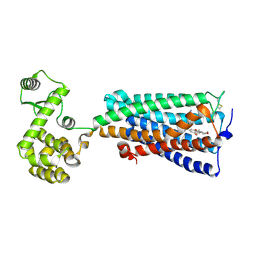 | | Crystal structure of an agonist bound GPCR | | Descriptor: | 7-[(6aR,9R,10aR)-1-Hydroxy-9-(hydroxymethyl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydro-6H-benzo[c]chromen-3-yl]- 7-methyloctanenitrile, Cannabinoid receptor 2,Endolysin | | Authors: | Li, X.T, Hua, T, Wu, L.J, Makriyannis, A, Wu, M, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
6KPF
 
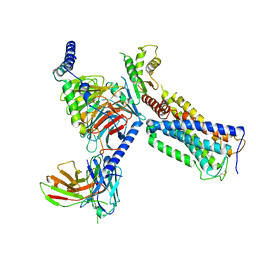 | | Cryo-EM structure of a class A GPCR with G protein complex | | Descriptor: | 7-[(6aR,9R,10aR)-1-Hydroxy-9-(hydroxymethyl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydro-6H-benzo[c]chromen-3-yl]- 7-methyloctanenitrile, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, X.T, Hua, T, Wu, L.J, Makriyannis, A, Shen, L, Wang, Y.X, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
5CMU
 
 | |
6KPG
 
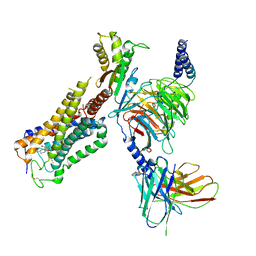 | | Cryo-EM structure of CB1-G protein complex | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hua, T, Li, X.T, Wu, L.J, Makriyannis, A, Wang, Y.X, Shen, L, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
5XRA
 
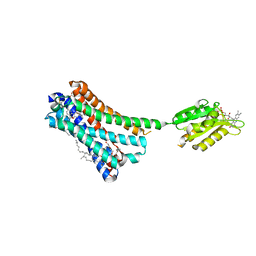 | | Crystal structure of the human CB1 in complex with agonist AM11542 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6aR,10aR)-3-(8-bromanyl-2-methyl-octan-2-yl)-6,6,9-trimethyl-6a,7,10,10a-tetrahydrobenzo[c]chromen-1-ol, CHOLESTEROL, ... | | Authors: | Hua, T, Vemuri, K, Nikas, P.S, Laprairie, R.B, Wu, Y, Qu, L, Pu, M, Korde, A, Shan, J, Ho, J.H, Han, G.W, Ding, K, Li, X, Liu, H, Hanson, M.A, Zhao, S, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2017-06-08 | | Release date: | 2017-07-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of agonist-bound human cannabinoid receptor CB1
Nature, 547, 2017
|
|
4N4H
 
 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.1K36me3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.1, ... | | Authors: | Li, Y, Ren, Y, Li, H. | | Deposit date: | 2013-10-08 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
5XR8
 
 | | Crystal structure of the human CB1 in complex with agonist AM841 | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, CHOLESTEROL, Cannabinoid receptor 1,Flavodoxin,Cannabinoid receptor 1, ... | | Authors: | Hua, T, Vemuri, K, Nikas, P.S, Laprairie, R.B, Wu, Y, Qu, L, Pu, M, Korde, A, Shan, J, Ho, J.H, Han, G.W, Ding, K, Li, X, Liu, H, Hanson, M.A, Zhao, S, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structures of agonist-bound human cannabinoid receptor CB1.
Nature, 547, 2017
|
|
4N4G
 
 | |
4N4I
 
 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.3K36me3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.3, ... | | Authors: | Li, Y, Ren, Y, Li, H. | | Deposit date: | 2013-10-08 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
5WYX
 
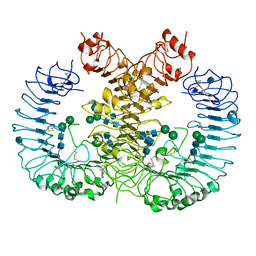 | | Crystal structure of human TLR8 in complex with CU-CPT8m | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(3-methylphenyl)pyrazolo[1,5-a]pyrimidine-3-carboxamide, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2017-01-16 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Small-molecule inhibition of TLR8 through stabilization of its resting state
Nat. Chem. Biol., 14, 2018
|
|
6L6H
 
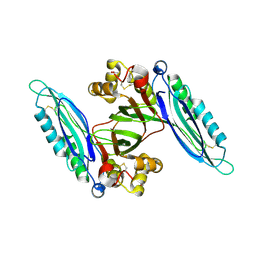 | | Crystal structure of Lpg0189 | | Descriptor: | ACETATE ION, GLYCEROL, Uncharacterized protein Lpg0189 | | Authors: | Ge, H, Chen, X. | | Deposit date: | 2019-10-29 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Crystal structure of a hypothetical T2SS effector Lpg0189 from Legionella pneumophila reveals a novel protein fold.
Biochem.Biophys.Res.Commun., 521, 2020
|
|
6L6G
 
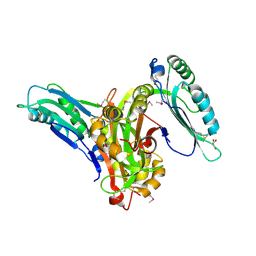 | | Crystal structure of SeMet_Lpg0189 | | Descriptor: | GLYCEROL, Uncharacterized protein Lpg0189 | | Authors: | Ge, H, Chen, X. | | Deposit date: | 2019-10-29 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of a hypothetical T2SS effector Lpg0189 from Legionella pneumophila reveals a novel protein fold.
Biochem.Biophys.Res.Commun., 521, 2020
|
|
5Z15
 
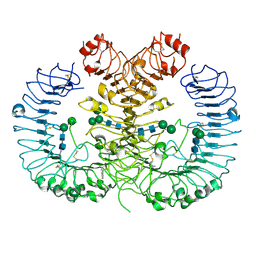 | | Crystal structure of human TLR8 in complex with CU-CPT9c | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(7-chloranylquinolin-4-yl)-2-methyl-phenol, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2017-12-25 | | Release date: | 2018-07-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inhibition of Toll-like Receptor 8 by Specifically Targeting a Unique Allosteric Site and Locking Its Resting State
To Be Published
|
|
6M4J
 
 | | SspA in complex with cysteine | | Descriptor: | CYSTEINE, PYRIDOXAL-5'-PHOSPHATE, SspA complex protein | | Authors: | Liu, L, Gao, H. | | Deposit date: | 2020-03-07 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of an l-Cysteine Desulfurase from an Ssp DNA Phosphorothioation System.
Mbio, 11, 2020
|
|
3WJ7
 
 | | Crystal structure of gox2253 | | Descriptor: | MERCURY (II) ION, Putative oxidoreductase | | Authors: | Yuan, Y.A, Yin, B. | | Deposit date: | 2013-10-05 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into substrate and coenzyme preference by SDR family protein Gox2253 from Gluconobater oxydans.
Proteins, 82, 2014
|
|
4BV6
 
 | | Refined crystal structure of the human Apoptosis inducing factor | | Descriptor: | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Martinez-Julvez, M, Herguedas, B, Hermoso, J.A, Ferreira, P, Villanueva, R, Medina, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into the Coenzyme Mediated Monomer-Dimer Transition of the Pro-Apoptotic Apoptosis Inducing Factor.
Biochemistry, 53, 2014
|
|
5C5T
 
 | |
7EZW
 
 | | Cyclic Peptide that Interacts with the eIF4E Capped-mRNA Binding Site | | Descriptor: | ALA-CYS-GLU-MET-GLY-PHE-PHE-GLN-ASP-CYS-GLY, Eukaryotic translation initiation factor 4E, SODIUM ION | | Authors: | Brown, C.J, Ng, S, Frosi, Y. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Development of a novel peptide aptamer that interacts with the eIF4E capped-mRNA binding site using peptide epitope linker evolution (PELE).
Rsc Chem Biol, 3, 2022
|
|
7F07
 
 | | Autonomous VH domain that interacts with eIF4E at the Capped mRNA Binding site. | | Descriptor: | Eukaryotic translation initiation factor 4E, VH domain (VH-DiFCAP-01) | | Authors: | Brown, C.J, Frosi, Y, Ng, S, Lin, Y.C. | | Deposit date: | 2021-06-03 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Development of a novel peptide aptamer that interacts with the eIF4E capped-mRNA binding site using peptide epitope linker evolution (PELE).
Rsc Chem Biol, 3, 2022
|
|
5UR1
 
 | | FGFR1 kinase domain complex with SN37333 in reversible binding mode | | Descriptor: | 3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-{1-[4-(dimethylamino)but-2-enoyl]piperidin-4-yl}-7-(phenylamino)-3,4-dihydropyrimido[4,5-d]pyrimidin-2(1H)-one, Fibroblast growth factor receptor 1 | | Authors: | Yosaatmadja, Y, Paik, W.-K, Smaill, J.B, Squire, C.J. | | Deposit date: | 2017-02-09 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2-Oxo-3, 4-dihydropyrimido[4, 5-d]pyrimidinyl derivatives as new irreversible pan fibroblast growth factor receptor (FGFR) inhibitors.
Eur J Med Chem, 135, 2017
|
|
6SJ7
 
 | |
7FEM
 
 | | SARS-CoV-2 B.1.1.7 S-ACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wen, Z.L, Zhu, Y, Sun, F. | | Deposit date: | 2021-07-21 | | Release date: | 2021-12-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure-based evidence for the enhanced transmissibility of the dominant SARS-CoV-2 B.1.1.7 variant (Alpha).
Cell Discov, 7, 2021
|
|
7FET
 
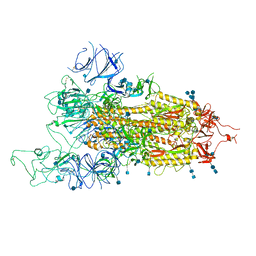 | | SARS-CoV-2 B.1.1.7 Spike Glycoprotein trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wen, Z.L, Zhu, Y, Sun, F. | | Deposit date: | 2021-07-21 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure-based evidence for the enhanced transmissibility of the dominant SARS-CoV-2 B.1.1.7 variant (Alpha).
Cell Discov, 7, 2021
|
|
4N0A
 
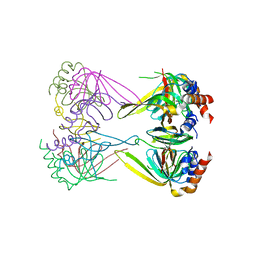 | | Crystal structure of Lsm2-3-Pat1C complex from Saccharomyces cerevisiae | | Descriptor: | DNA topoisomerase 2-associated protein PAT1, U6 snRNA-associated Sm-like protein LSm2, U6 snRNA-associated Sm-like protein LSm3 | | Authors: | Wu, D.H. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Lsm2 and Lsm3 bridge the interaction of the Lsm1-7 complex with Pat1 for decapping activation
Cell Res., 24, 2014
|
|
5FEC
 
 | |
