1SJ3
 
 | | Hepatitis Delta Virus Gemonic Ribozyme Precursor, with Mg2+ Bound | | Descriptor: | MAGNESIUM ION, precursor form of the Hepatitis Delta virus ribozyme, small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H, Doudna, J.A. | | Deposit date: | 2004-03-02 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme
catalysis
Nature, 429, 2004
|
|
1VBY
 
 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutaion, and Mn2+ bound | | Descriptor: | Hepatitis Delta virus ribozyme, MANGANESE (II) ION, SODIUM ION, ... | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
1VC0
 
 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutaion, in Imidazole and Sr2+ solution | | Descriptor: | Hepatitis Delta virus ribozyme, STRONTIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
1VC6
 
 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Product with C75U Mutaion, cleaved in Imidazole and Mg2+ solutions | | Descriptor: | Hepatitis Delta virus ribozyme, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-04 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
1VBX
 
 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutaion, in EDTA solution | | Descriptor: | Hepatitis Delta virus ribozyme, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
1VC5
 
 | | Crystal Structure of the Wild Type Hepatitis Delta Virus Gemonic Ribozyme Precursor, in EDTA solution | | Descriptor: | Hepatitis Delta virus ribozyme, SODIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-04 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
1VBZ
 
 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutaion, in Ba2+ solution | | Descriptor: | BARIUM ION, Hepatitis Delta virus ribozyme, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
1SJ4
 
 | | Crystal structure of a C75U mutant Hepatitis Delta Virus ribozyme precursor, in Cu2+ solution | | Descriptor: | precursor form of the Hepatitis Delta virus ribozyme, small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H, Doudna, J.A. | | Deposit date: | 2004-03-02 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme
catalysis
Nature, 429, 2004
|
|
3LQX
 
 | | SRP ribonucleoprotein core complexed with cobalt hexammine | | Descriptor: | CHLORIDE ION, COBALT HEXAMMINE(III), POTASSIUM ION, ... | | Authors: | Batey, R.T. | | Deposit date: | 2010-02-10 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and Energetic Analysis of Metal Ions Essential to SRP Signal Recognition Domain Assembly
Biochemistry, 41, 2002
|
|
8DC2
 
 | | Cryo-EM structure of CasLambda (Cas12l) bound to crRNA and DNA | | Descriptor: | CasLambda, DNA NTS, DNA TS, ... | | Authors: | Al-Shayeb, B, Skopintsev, P, Soczek, K, Doudna, J. | | Deposit date: | 2022-06-15 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Diverse virus-encoded CRISPR-Cas systems include streamlined genome editors.
Cell, 185, 2022
|
|
7WB0
 
 | |
7WAZ
 
 | |
7WAY
 
 | |
7WB1
 
 | |
8FYA
 
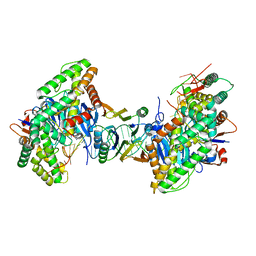 | | Cryo-EM structure of Cas1:Cas2-DEDDh:PAM-containing prespacer complex | | Descriptor: | Cas1, Cas2-DEDDh, DNA (28-MER), ... | | Authors: | Skopintsev, P, Tuck, O.T, Soczek, K.M, Doudna, J. | | Deposit date: | 2023-01-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Genome expansion by a CRISPR trimmer-integrase.
Nature, 618, 2023
|
|
8FYB
 
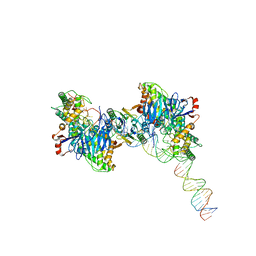 | | Cryo-EM structure of Cas1:Cas2-DEDDh:half-site integration complex | | Descriptor: | Cas1, Cas2-DEDDh, DNA (17-MER), ... | | Authors: | Skopintsev, P, Tuck, O.T, Soczek, K.M, Doudna, J. | | Deposit date: | 2023-01-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Genome expansion by a CRISPR trimmer-integrase.
Nature, 618, 2023
|
|
8FYC
 
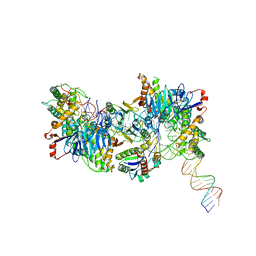 | | Cryo-EM structure of Cas1:Cas2-DEDDh:half-site integration complex linear CRISPR repeat conformation | | Descriptor: | Cas1, Cas2-DEDDh, DEDDh, ... | | Authors: | Skopintsev, P, Tuck, O.T, Soczek, K.M, Doudna, J. | | Deposit date: | 2023-01-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Genome expansion by a CRISPR trimmer-integrase.
Nature, 618, 2023
|
|
8FY9
 
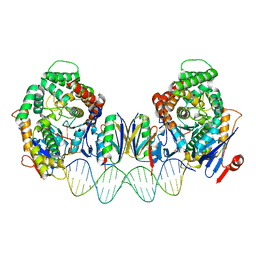 | | Cryo-EM structure of Cas1:Cas2-DEDDh:PAM-deficient prespacer complex | | Descriptor: | Cas1, Cas2-DEDDh, DNA (28-MER) | | Authors: | Skopintsev, P, Tuck, O.T, Soczek, K.M, Doudna, J. | | Deposit date: | 2023-01-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Genome expansion by a CRISPR trimmer-integrase.
Nature, 618, 2023
|
|
8FYD
 
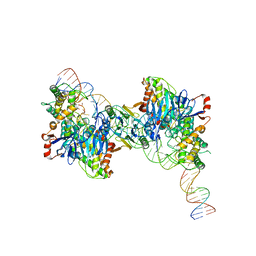 | | Cryo-EM structure of Cas1:Cas2-DEDDh:half-site integration complex bent CRISPR repeat conformation | | Descriptor: | Cas1, Cas2-DEDDh, DNA (13-MER), ... | | Authors: | Skopintsev, P, Tuck, O.T, Soczek, K.M, Doudna, J. | | Deposit date: | 2023-01-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Genome expansion by a CRISPR trimmer-integrase.
Nature, 618, 2023
|
|
1HR2
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A MUTANT P4-P6 DOMAIN (DELC209) OF TETRAHYMENA THEMOPHILA GROUP I INTRON. | | Descriptor: | MAGNESIUM ION, P4-P6 DELC209 MUTANT RNA RIBOZYME DOMAIN | | Authors: | Juneau, K, Podell, E.R, Harrington, D.J, Cech, T.R. | | Deposit date: | 2000-12-20 | | Release date: | 2001-04-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA--solvent interactions.
Structure, 9, 2001
|
|
7LYS
 
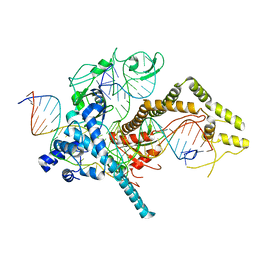 | | Cryo-EM structure of CasPhi-2 (Cas12j) bound to crRNA and DNA | | Descriptor: | CasPhi-2, NTS-DNA, TS-DNA, ... | | Authors: | Pausch, P, Soczek, K, Nogales, E, Doudna, J. | | Deposit date: | 2021-03-08 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | DNA interference states of the hypercompact CRISPR-Cas Phi effector.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LYT
 
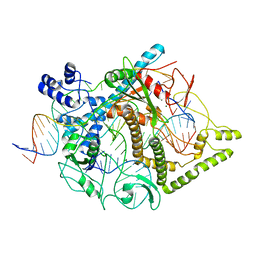 | | Cryo-EM structure of CasPhi-2 (Cas12j) bound to crRNA and Phosphorothioate-DNA | | Descriptor: | CasPhi, MAGNESIUM ION, NTS-DNA*, ... | | Authors: | Pausch, P, Soczek, K, Nogales, E, Doudna, J. | | Deposit date: | 2021-03-08 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | DNA interference states of the hypercompact CRISPR-Cas Phi effector.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M5O
 
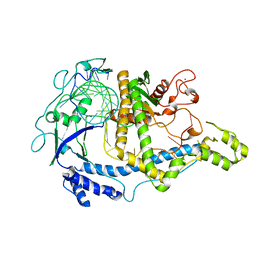 | | Cryo-EM structure of CasPhi-2 (Cas12j) bound to crRNA | | Descriptor: | CasPhi, ZINC ION, crRNA | | Authors: | Pausch, P, Soczek, K, Nogales, E, Doudna, J. | | Deposit date: | 2021-03-24 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | DNA interference states of the hypercompact CRISPR-Cas Phi effector.
Nat.Struct.Mol.Biol., 28, 2021
|
|
280D
 
 | |
3GOD
 
 | |
