6FSN
 
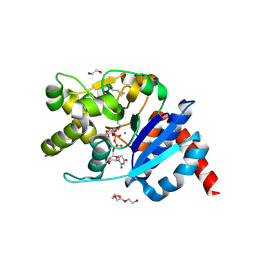 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum in complex with UDP-glucose (conformation 1) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Roversi, P, Le Cornu, J.D, Hill, J, Alonzi, D.S, Zitzmann, N. | | Deposit date: | 2018-02-19 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Crystal polymorphism in fragment-based lead discovery of ligands of the catalytic domain of UGGT, the glycoprotein folding quality control checkpoint.
Front Mol Biosci, 2022
|
|
3W51
 
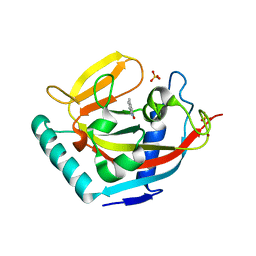 | | Tankyrase in complex with 2-hydroxy-4-methylquinoline | | Descriptor: | 4-methylquinolin-2-ol, SULFATE ION, Tankyrase-2, ... | | Authors: | Jansson, A.E, Larsson, E.A, Nordlund, P.L. | | Deposit date: | 2013-01-18 | | Release date: | 2013-07-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-based ligand design of novel potent inhibitors of tankyrases.
J.Med.Chem., 56, 2013
|
|
4EJN
 
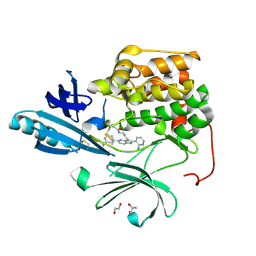 | | Crystal structure of autoinhibited form of AKT1 in complex with N-(4-(5-(3-acetamidophenyl)-2-(2-aminopyridin-3-yl)-3H-imidazo[4,5-b]pyridin-3-yl)benzyl)-3-fluorobenzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-BUTANOL, N-(4-{5-[3-(acetylamino)phenyl]-2-(2-aminopyridin-3-yl)-3H-imidazo[4,5-b]pyridin-3-yl}benzyl)-3-fluorobenzamide, ... | | Authors: | Eathiraj, S. | | Deposit date: | 2012-04-06 | | Release date: | 2012-05-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery and optimization of a series of 3-(3-phenyl-3H-imidazo[4,5-b]pyridin-2-yl)pyridin-2-amines: orally bioavailable, selective, and potent ATP-independent Akt inhibitors.
J.Med.Chem., 55, 2012
|
|
1PFZ
 
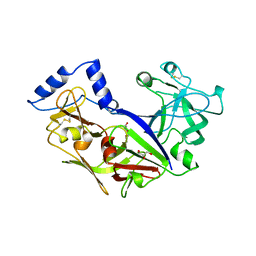 | | PROPLASMEPSIN II FROM PLASMODIUM FALCIPARUM | | Descriptor: | GLYCEROL, PROPLASMEPSIN II | | Authors: | Bernstein, N.K, Cherney, M.M, Loetscher, H, Ridley, R.G, James, M.N.G. | | Deposit date: | 1998-07-07 | | Release date: | 1999-01-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the novel aspartic proteinase zymogen proplasmepsin II from plasmodium falciparum.
Nat.Struct.Biol., 6, 1999
|
|
4J22
 
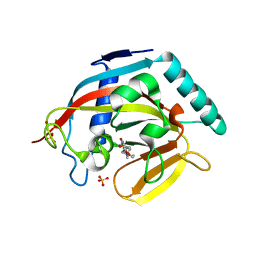 | | Tankyrase 2 in complex with 3-chloro-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)-N-[2-(morpholin-4-yl)ethyl]benzamide | | Descriptor: | 3-chloro-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)-N-[2-(morpholin-4-yl)ethyl]benzamide, SULFATE ION, Tankyrase-2, ... | | Authors: | Jansson, A.E, Larsson, E.A, Nordlund, P.L. | | Deposit date: | 2013-02-04 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Fragment-based ligand design of novel potent inhibitors of tankyrases.
J.Med.Chem., 56, 2013
|
|
4IUE
 
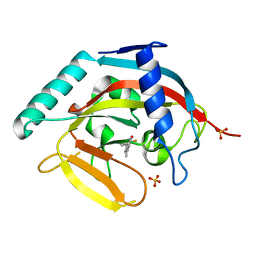 | | Tankyrase in complex with 7-(2-fluorophenyl)-4-methyl-1,2-dihydroquinolin-2-one | | Descriptor: | 7-(2-fluorophenyl)-4-methylquinolin-2(1H)-one, SULFATE ION, Tankyrase-2, ... | | Authors: | Jansson, A.E, Larsson, E.A, Nordlund, P.L. | | Deposit date: | 2013-01-21 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Fragment-based ligand design of novel potent inhibitors of tankyrases.
J.Med.Chem., 56, 2013
|
|
4J3M
 
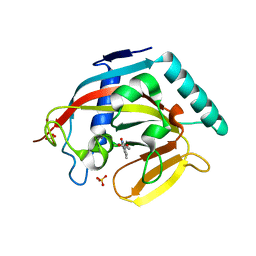 | | Tankyrase 2 in complex with 3-chloro-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)benzoic acid | | Descriptor: | 3-chloro-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)benzoic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Jansson, A.E, Larsson, E.A, Nordlund, P.L. | | Deposit date: | 2013-02-06 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-based ligand design of novel potent inhibitors of tankyrases.
J.Med.Chem., 56, 2013
|
|
4J1Z
 
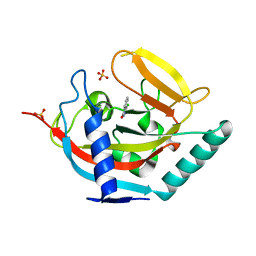 | |
4J21
 
 | | Tankyrase 2 in complex with 7-(4-amino-2-chlorophenyl)-4-methylquinolin-2(1H)-one | | Descriptor: | 7-(4-amino-2-chlorophenyl)-4-methylquinolin-2(1H)-one, SULFATE ION, Tankyrase-2, ... | | Authors: | Jansson, A.E, Larsson, E.A, Nordlund, P.L. | | Deposit date: | 2013-02-04 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Fragment-based ligand design of novel potent inhibitors of tankyrases.
J.Med.Chem., 56, 2013
|
|
4J3L
 
 | | Tankyrase 2 in complex with 3-chloro-N-(2-methoxyethyl)-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)benzamide | | Descriptor: | 3-chloro-N-(2-methoxyethyl)-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)benzamide, SULFATE ION, Tankyrase-2, ... | | Authors: | Jansson, A.E, Larsson, E.A, Nordlund, P.L. | | Deposit date: | 2013-02-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Fragment-based ligand design of novel potent inhibitors of tankyrases.
J.Med.Chem., 56, 2013
|
|
8B22
 
 | |
8B23
 
 | |
8B21
 
 | | Time-resolved structure of K+-dependent Na+-PPase from Thermotoga maritima 0-60-seconds post reaction initiation with Na+ | | Descriptor: | DI(HYDROXYETHYL)ETHER, DODECYL-BETA-D-MALTOSIDE, K(+)-stimulated pyrophosphate-energized sodium pump, ... | | Authors: | Strauss, J, Vidilaseris, K, Goldman, A. | | Deposit date: | 2022-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
8B24
 
 | | Time-resolved structure of K+-dependent Na+-PPase from Thermotoga maritima 3600-seconds post reaction initiation with Na+ | | Descriptor: | DIPHOSPHATE, K(+)-stimulated pyrophosphate-energized sodium pump, MAGNESIUM ION, ... | | Authors: | Strauss, J, Vidilaseris, K, Goldman, A. | | Deposit date: | 2022-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (4.53 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
6Z0E
 
 | |
6Z0X
 
 | |
6Z0Y
 
 | |
6YSK
 
 | | 1-phenylpyrroles and 1-enylpyrrolidines as inhibitors of Notum | | Descriptor: | (3~{S})-1-[4-chloranyl-3-(trifluoromethyl)phenyl]pyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2020-04-22 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6TV4
 
 | | CFF-Notum complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CAFFEINE, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-01-08 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Caffeine inhibits Notum activity by binding at the catalytic pocket.
Commun Biol, 3, 2020
|
|
6TUZ
 
 | | Theophylline-Notum complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-01-08 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Caffeine inhibits Notum activity by binding at the catalytic pocket.
Commun Biol, 3, 2020
|
|
7PK3
 
 | | Notum_ARUK3001185 | | Descriptor: | 1-[2,4-bis(chloranyl)-3-(trifluoromethyl)phenyl]-1,2,3-triazole, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Vecchia, L, Hillier, J, Zhao, Y, Fish, P, Jones, E.Y. | | Deposit date: | 2021-08-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Design of a Potent, Selective, and Brain-Penetrant Inhibitor of Wnt-Deactivating Enzyme Notum by Optimization of a Crystallographic Fragment Hit.
J.Med.Chem., 65, 2022
|
|
6ZWT
 
 | | Crystal structure of DNA-binding domain of OmpR of two-component system of Acinetobacter baumannii | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Narwal, M, Lucchini, V, Trebosc, V, Gartenmann, S, Pieren, M, Kemmer, C, Kammerer, R.A. | | Deposit date: | 2020-07-28 | | Release date: | 2021-08-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeting virulence regulation to disarm Acinetobacter baumannii pathogenesis.
Virulence, 13, 2022
|
|
6ZVL
 
 | | ARUK3000263 complex with Notum | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[4-chloranyl-3-(trifluoromethyl)phenyl]-3~{H}-1,3,4-oxadiazol-2-one, ... | | Authors: | Zhao, Y, Ruza, R. | | Deposit date: | 2020-07-24 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 5-Phenyl-1,3,4-oxadiazol-2(3 H )-ones Are Potent Inhibitors of Notum Carboxylesterase Activity Identified by the Optimization of a Crystallographic Fragment Screening Hit.
J.Med.Chem., 63, 2020
|
|
6ZUV
 
 | | Notum fragment 286 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-07-23 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | 5-Phenyl-1,3,4-oxadiazol-2(3 H )-ones Are Potent Inhibitors of Notum Carboxylesterase Activity Identified by the Optimization of a Crystallographic Fragment Screening Hit.
J.Med.Chem., 63, 2020
|
|
6TR5
 
 | | Melatonin-Notum complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.509 Å) | | Cite: | Structural characterization of melatonin as an inhibitor of the Wnt deacylase Notum.
J. Pineal Res., 68, 2020
|
|
