8DKB
 
 | |
8GOU
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH003 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH003 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
8GPY
 
 | | Crystal structure of Omicron BA.4/5 RBD in complex with a neutralizing antibody scFv | | Descriptor: | Spike protein S1, scFv | | Authors: | Gao, Y.X, Song, Z.D, Wang, W.M, Guo, Y. | | Deposit date: | 2022-08-27 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
4YT6
 
 | |
4YT7
 
 | |
7RX2
 
 | |
7RXH
 
 | |
7RXA
 
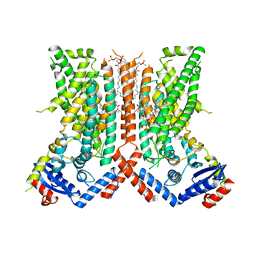 | | afTMEM16 DE/AA mutant in C14 lipid nanodiscs in the presence of Ca2+ | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, afTMEM16 lipid scramblase | | Authors: | Feng, Z, Accardi, A. | | Deposit date: | 2021-08-22 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | TMEM16 scramblases thin the membrane to enable lipid scrambling.
Nat Commun, 13, 2022
|
|
7RX3
 
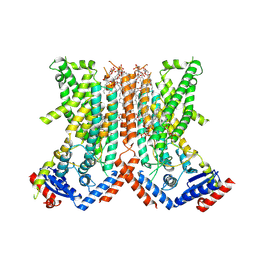 | |
7RXG
 
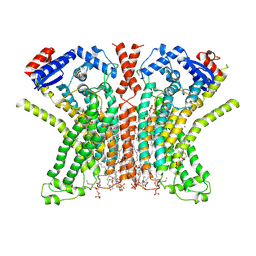 | |
7RWJ
 
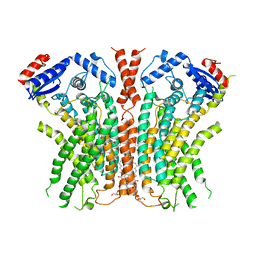 | |
7RXB
 
 | | afTMEM16 lipid scramblase in C18 lipid nanodiscs in the absence of Ca2+ | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, afTMEM16 lipid scramblase | | Authors: | Feng, Z, Accardi, A. | | Deposit date: | 2021-08-22 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | TMEM16 scramblases thin the membrane to enable lipid scrambling.
Nat Commun, 13, 2022
|
|
9B4J
 
 | | Filament of D-TLKIVWS, a D-peptide that disaggregates Alzheimer's Paired Helical Filaments, determined by Cryo-EM | | Descriptor: | DTH-DLE-DLY-DIL-DVA-DTR-DSN | | Authors: | Hou, K, Ge, P, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2024-03-20 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | How short peptides disassemble tau fibrils in Alzheimer's disease
Nature, 2025
|
|
9B4K
 
 | | Filament of D-TLKIVWR, a D-peptide that disaggregates Alzheimer's Paired Helical Filaments, determined by Cryo-EM | | Descriptor: | DTH-DLE-DLY-DIL-DVA-DTR-DAR | | Authors: | Hou, K, Ge, P, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2024-03-20 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | How short peptides disassemble tau fibrils in Alzheimer's disease
Nature, 2025
|
|
9B4O
 
 | | Alzheimer's Tau Paired Helical Filaments, determined by CryoEM, before addition of D-peptide disaggregants | | Descriptor: | Microtubule-associated protein tau | | Authors: | Hou, K, Ge, P, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2024-03-21 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | How short peptides disassemble tau fibrils in Alzheimer's disease
Nature, 2025
|
|
9B4I
 
 | | Filament of D-TLKIVWI, a D-peptide that disaggregates Alzheimer's Paired Helical Filaments, determined by Cryo-EM | | Descriptor: | DTH-DLE-DLY-DIL-DVA-DTR-DIL | | Authors: | Hou, K, Ge, P, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2024-03-20 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | How short peptides disassemble tau fibrils in Alzheimer's disease
Nature, 2025
|
|
9B4N
 
 | | Filament of Tau in complex with D-TLKIVWR, a D-peptide that disaggregates Alzheimer's Paired Helical Filaments, determined by Cryo-EM | | Descriptor: | Microtubule-associated protein tau | | Authors: | Hou, K, Ge, P, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2024-03-21 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | How short peptides disassemble tau fibrils in Alzheimer's disease
Nature, 2025
|
|
9B4M
 
 | | Filament of Tau in complex with D-TLKIVWS, a D-peptide that disaggregates Alzheimer's Paired Helical Filaments, determined by Cryo-EM | | Descriptor: | Microtubule-associated protein tau | | Authors: | Hou, K, Ge, P, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2024-03-20 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | How short peptides disassemble tau fibrils in Alzheimer's disease
Nature, 2025
|
|
9B4L
 
 | | Filament of Tau in complex with D-TLKIVWI, a D-peptide that disaggregates Alzheimer's Paired Helical Filaments, determined by Cryo-EM | | Descriptor: | DTH-DLE-DLY-DIL-DVA-DTR-DIL, Microtubule-associated protein tau, {[-(BIS-CARBOXYMETHYL-AMINO)-ETHYL]-CARBOXYMETHYL-AMINO}-ACETIC ACID | | Authors: | Hou, K, Ge, P, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2024-03-20 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | How short peptides disassemble tau fibrils in Alzheimer's disease
Nature, 2025
|
|
3NEW
 
 | | p38-alpha complexed with Compound 10 | | Descriptor: | 4-(trifluoromethyl)-3-[3-(trifluoromethyl)phenyl]-1,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, Mitogen-activated protein kinase 14 | | Authors: | Goedken, E.R, Comess, K.M, Sun, C, Argiriadi, M, Jia, Y, Quinn, C.M, Banach, D.L, Marcotte, D, Borhani, D. | | Deposit date: | 2010-06-09 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and Characterization of Non-ATP Site Inhibitors of the Mitogen Activated Protein (MAP) Kinases.
Acs Chem.Biol., 6, 2011
|
|
3O17
 
 | | Crystal Structure of JNK1-alpha1 isoform | | Descriptor: | C-Jun-amino-terminal kinase-interacting protein 1, JIP1, 10MER PEPTIDE, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2010-07-20 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of JNK1-alpha1 isoform
TO BE PUBLISHED
|
|
8K04
 
 | | CryoEM structure of a 2,3-hydroxycinnamic acid 1,2-dioxygenase MhpB in apo form | | Descriptor: | 2,3-dihydroxyphenylpropionate/2,3-dihydroxicinnamic acid 1,2-dioxygenase | | Authors: | Jiang, W.X, Cheng, X.Q, Ma, L.X, Xing, Q. | | Deposit date: | 2023-07-07 | | Release date: | 2024-07-10 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural and catalytic insights into MhpB: A dioxygenase enzyme for degrading catecholic pollutants.
J Hazard Mater, 488, 2025
|
|
4XIC
 
 | | ANTPHD WITH 15BP di-thioate modified DNA DUPLEX | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*GP*CP*(C2S)P*AP*TP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), ... | | Authors: | White, M.A, Zandarashvili, L, Iwahara, J. | | Deposit date: | 2015-01-06 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Entropic Enhancement of Protein-DNA Affinity by Oxygen-to-Sulfur Substitution in DNA Phosphate.
Biophys.J., 109, 2015
|
|
5VGU
 
 | |
4X9J
 
 | | EGR-1 with Doubly Methylated DNA | | Descriptor: | DNA (5'-D(*AP*GP*(5CM)P*GP*TP*GP*GP*GP*(5CM)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*(5CM)P*GP*C)-3'), Early growth response protein 1, ... | | Authors: | White, M.A, Zandarashvili, L, Iwahara, J. | | Deposit date: | 2014-12-11 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.412 Å) | | Cite: | Structural impact of complete CpG methylation within target DNA on specific complex formation of the inducible transcription factor Egr-1.
Febs Lett., 589, 2015
|
|
