5ZNL
 
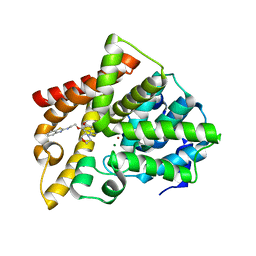 | | Crystal structure of PDE10A catalytic domain complexed with LHB-6 | | Descriptor: | MAGNESIUM ION, N-[2-(7-methoxy-4-morpholin-4-yl-quinazolin-6-yl)oxyethyl]-1,3-benzothiazol-2-amine, ZINC ION, ... | | Authors: | Li, Z, Huang, Y, Zhan, C.G, Luo, H.B. | | Deposit date: | 2018-04-09 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Absolute Binding Free Energy Calculation and Design of a Subnanomolar Inhibitor of Phosphodiesterase-10.
J. Med. Chem., 62, 2019
|
|
7UL1
 
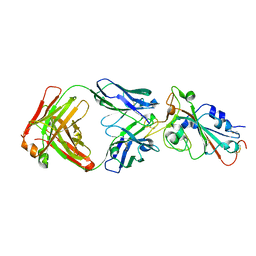 | | Crystal structure of SARS-CoV-2 RBD in complex with the neutralizing IGHV3-53-encoded antibody EH3 isolated from a nonvaccinated pediatric patient | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of EH3, Light chain of EH3, ... | | Authors: | Chen, Y, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2022-04-03 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular basis for antiviral activity of two pediatric neutralizing antibodies targeting SARS-CoV-2 Spike RBD.
Iscience, 26, 2023
|
|
7UL0
 
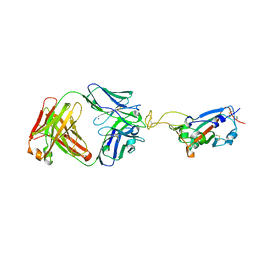 | | Crystal structure of SARS-CoV-2 RBD in complex with the ridge-binding nAb EH8 isolated from a nonvaccinated pediatric patient | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of EH8, ... | | Authors: | Chen, Y, Tolbert, W.D, Bai, X, Pazgier, M. | | Deposit date: | 2022-04-03 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular basis for antiviral activity of two pediatric neutralizing antibodies targeting SARS-CoV-2 Spike RBD.
Iscience, 26, 2023
|
|
6OQU
 
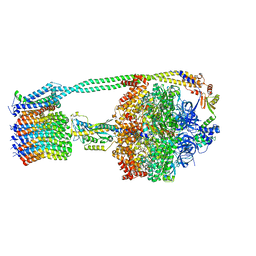 | | E. coli ATP synthase State 1d | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
6OQR
 
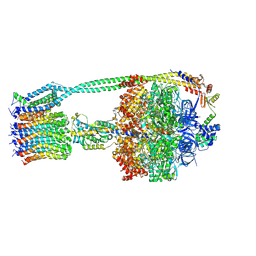 | | E. coli ATP Synthase ADP State 1a | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Walshe, J.L, Sobti, M. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-03 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
6A7A
 
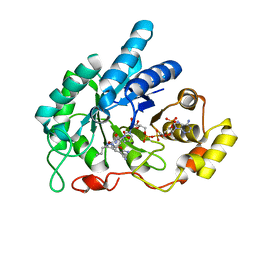 | | AKR1C1 complexed with new inhibitor with novel scaffold | | Descriptor: | (4R)-6-amino-4-(4-hydroxy-3-methoxy-5-nitrophenyl)-3-propyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X, Zhao, Y, Zhang, H, Chen, Y. | | Deposit date: | 2018-07-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
6KK0
 
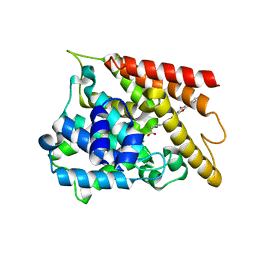 | | Crystal structure of PDE4D catalytic domain complexed with compound 4e | | Descriptor: | 7-[8-methoxy-2,2-dimethyl-7-(3-methylbut-2-enyl)-5-oxidanyl-6-oxidanylidene-pyrano[3,2-b]xanthen-9-yl]oxyheptanoic acid, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huang, Y.-Y, He, X, Luo, H.-B. | | Deposit date: | 2019-07-23 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.20008755 Å) | | Cite: | Discovery and Optimization of alpha-Mangostin Derivatives as Novel PDE4 Inhibitors for the Treatment of Vascular Dementia.
J.Med.Chem., 63, 2020
|
|
5ZI1
 
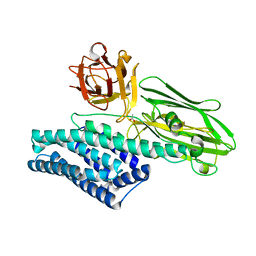 | |
6KJZ
 
 | | Crystal structure of PDE4D catalytic domain complexed with compound 1 | | Descriptor: | 8-methoxy-2,2-dimethyl-7-(3-methylbut-2-enyl)-5,9-bis(oxidanyl)pyrano[3,2-b]xanthen-6-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huang, Y.-Y, He, X, Luo, H.-B. | | Deposit date: | 2019-07-23 | | Release date: | 2020-03-18 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.200001 Å) | | Cite: | Discovery and Optimization of alpha-Mangostin Derivatives as Novel PDE4 Inhibitors for the Treatment of Vascular Dementia.
J.Med.Chem., 63, 2020
|
|
7SHF
 
 | | Cryo-EM structure of GPR158 coupled to the RGS7-Gbeta5 complex | | Descriptor: | (2S)-1-{[(S)-hydroxy{[(1s,2R,3R,4R,5S,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5E,8E,11E,14E)-icosa-5,8,11,14-tetraenoate, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, ... | | Authors: | Patil, D.N, Singh, S, Singh, A.K, Martemyanov, K.A. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-01 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of human GPR158 receptor coupled to the RGS7-G beta 5 signaling complex.
Science, 375, 2022
|
|
7SHE
 
 | | Cryo-EM structure of human GPR158 | | Descriptor: | (2S)-1-{[(S)-hydroxy{[(1s,2R,3R,4R,5S,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5E,8E,11E,14E)-icosa-5,8,11,14-tetraenoate, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, ... | | Authors: | Patil, D.N, Singh, S, Singh, A.K, Martemyanov, K.A. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-01 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of human GPR158 receptor coupled to the RGS7-G beta 5 signaling complex.
Science, 375, 2022
|
|
7XI3
 
 | | Crystal Structure of the NPAS4-ARNT2 heterodimer in complex with DNA | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator 2, DNA (5'-D(P*CP*CP*AP*TP*CP*AP*CP*TP*CP*AP*CP*GP*AP*CP*CP*T)-3'), DNA (5'-D(P*GP*GP*AP*GP*GP*TP*CP*GP*TP*GP*AP*GP*TP*GP*AP*T)-3'), ... | | Authors: | Sun, X.N, Jing, L.Q, Li, F.W, Wu, D.L. | | Deposit date: | 2022-04-11 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.274 Å) | | Cite: | Structures of NPAS4-ARNT and NPAS4-ARNT2 heterodimers reveal new dimerization modalities in the bHLH-PAS transcription factor family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TJ6
 
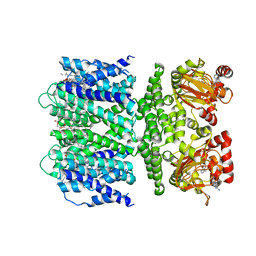 | | SthK open state, cAMP-bound in the presence of POPA | | Descriptor: | (2R)-1-(hexadecanoyloxy)-3-(phosphonooxy)propan-2-yl (9Z)-octadec-9-enoate, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Putative transcriptional regulator, ... | | Authors: | Schmidpeter, P.A, Nimigean, C.M. | | Deposit date: | 2022-01-14 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Anionic lipids unlock the gates of select ion channels in the pacemaker family.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TJ5
 
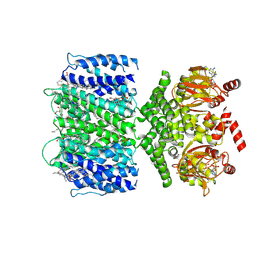 | | SthK closed state, cAMP-bound in the presence of POPA | | Descriptor: | (2R)-1-(hexadecanoyloxy)-3-(phosphonooxy)propan-2-yl (9Z)-octadec-9-enoate, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Putative transcriptional regulator, ... | | Authors: | Schmidpeter, P.A, Nimigean, C.M. | | Deposit date: | 2022-01-14 | | Release date: | 2022-10-26 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Anionic lipids unlock the gates of select ion channels in the pacemaker family.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TKT
 
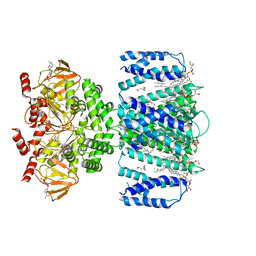 | | SthK closed state, cAMP-bound in the presence of detergent | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Putative transcriptional regulator, ... | | Authors: | Rheinberger, J, Schmidpeter, P.A, Nimigean, C.M. | | Deposit date: | 2022-01-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Anionic lipids unlock the gates of select ion channels in the pacemaker family.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7XI4
 
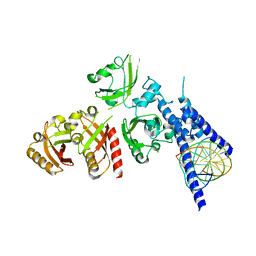 | | Crystal Structure of the NPAS4-ARNT heterodimer in complex with DNA | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, DNA (5'-D(*GP*GP*AP*GP*GP*TP*CP*GP*TP*GP*AP*GP*TP*GP*AP*T)-3'), DNA (5'-D(P*CP*CP*AP*TP*CP*AP*CP*TP*CP*AP*CP*GP*AP*CP*CP*T)-3'), ... | | Authors: | Sun, X.N, Jing, L.Q, Li, F.W, Wu, D.L. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.707 Å) | | Cite: | Structures of NPAS4-ARNT and NPAS4-ARNT2 heterodimers reveal new dimerization modalities in the bHLH-PAS transcription factor family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XHV
 
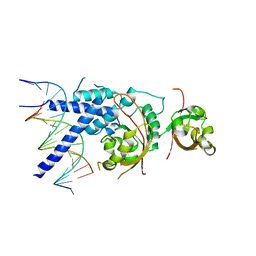 | | Crystal Structure of the NPAS4-ARNT heterodimer in complex with DNA | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, DNA (5'-D(P*CP*CP*AP*TP*CP*AP*CP*TP*CP*AP*CP*GP*AP*CP*CP*T)-3'), DNA (5'-D(P*GP*GP*AP*GP*GP*TP*CP*GP*TP*GP*AP*GP*TP*GP*AP*T)-3'), ... | | Authors: | Sun, X.N, Jing, L.Q, Li, F.W, Wu, D.L. | | Deposit date: | 2022-04-10 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.996 Å) | | Cite: | Structures of NPAS4-ARNT and NPAS4-ARNT2 heterodimers reveal new dimerization modalities in the bHLH-PAS transcription factor family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7U4T
 
 | | Human V-ATPase in state 2 with SidK and mEAK-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, MTOR-associated protein MEAK7, ... | | Authors: | Wang, L, Fu, T.M. | | Deposit date: | 2022-03-01 | | Release date: | 2022-08-24 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Identification of mEAK-7 as a human V-ATPase regulator via cryo-EM data mining.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8ZCS
 
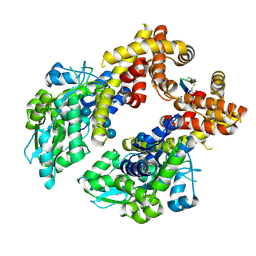 | | Crystal structure of the MBP-MCL1 complex with highly selective and potent Cyclic peptide inhibitor | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, TYR-LEU-LEU-PHE-TRP-ARG-ASP-GLU-LEU-ILE-LEU-LEU-CCJ-NH2, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Li, F.W. | | Deposit date: | 2024-04-30 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | De novo discovery of a molecular glue-like macrocyclic peptide that induces MCL1 homodimerization.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
6WNR
 
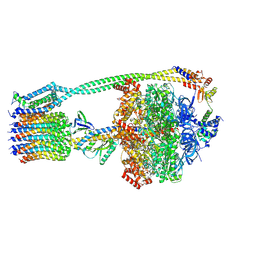 | | E. coli ATP synthase State 3b | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-03 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
6WNQ
 
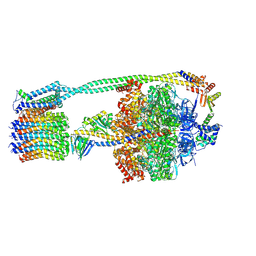 | | E. coli ATP Synthase State 2a | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-03 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
5K76
 
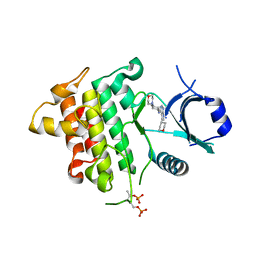 | | IRAK4 in complex with Compound 28 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N}-(4-morpholin-4-ylcyclohexyl)-5-(oxan-4-yl)-7~{H}-pyrrolo[2,3-d]pyrimidin-4-amine | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-05-25 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Discovery and Optimization of Pyrrolopyrimidine Inhibitors of Interleukin-1 Receptor Associated Kinase 4 (IRAK4) for the Treatment of Mutant MYD88L265P Diffuse Large B-Cell Lymphoma.
J. Med. Chem., 60, 2017
|
|
5K7I
 
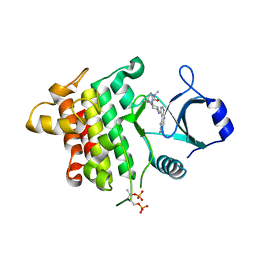 | | IRAK4 in complex with AZ3864 | | Descriptor: | (3~{a}~{R},7~{a}~{S})-1-methyl-5-[4-[[5-(oxan-4-yl)-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl]amino]cyclohexyl]-3,3~{a},4,6,7,7~{a}-hexahydropyrrolo[3,2-c]pyridin-2-one, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-05-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery and Optimization of Pyrrolopyrimidine Inhibitors of Interleukin-1 Receptor Associated Kinase 4 (IRAK4) for the Treatment of Mutant MYD88L265P Diffuse Large B-Cell Lymphoma.
J. Med. Chem., 60, 2017
|
|
6M4U
 
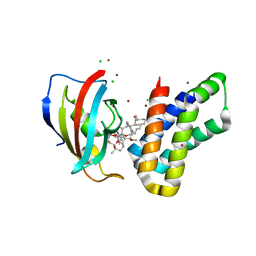 | | Crystal structure of FKBP-FRB T2098L mutant in complex with rapamycin | | Descriptor: | CHLORIDE ION, GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Kikuchi, M, Wu, D, Inoue, T, Umehara, T. | | Deposit date: | 2020-03-09 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational design and implementation of a chemically inducible heterotrimerization system.
Nat.Methods, 17, 2020
|
|
5K75
 
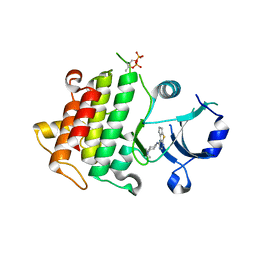 | | IRAK4 in complex with Compound 1 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, SULFATE ION, ~{N}1-(7,8-dihydro-6~{H}-cyclopenta[2,3]thieno[2,4-~{c}]pyrimidin-1-yl)-~{N}4,~{N}4-dimethyl-cyclohexane-1,4-diamine | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-05-25 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery and Optimization of Pyrrolopyrimidine Inhibitors of Interleukin-1 Receptor Associated Kinase 4 (IRAK4) for the Treatment of Mutant MYD88L265P Diffuse Large B-Cell Lymphoma.
J. Med. Chem., 60, 2017
|
|
