5AXI
 
 | | Crystal structure of Cbl-b TKB domain in complex with Cblin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cblin, ... | | Authors: | Ohno, A, Maita, N, Ochi, A, Nakao, R, Nikawa, T. | | Deposit date: | 2015-07-29 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the TKB domain of ubiquitin ligase Cbl-b complexed with its small inhibitory peptide, Cblin
Arch.Biochem.Biophys., 594, 2016
|
|
6XAR
 
 | |
5O76
 
 | | Structure of phosphoY371 c-CBL in complex with ZAP70-peptide and UbV.pCBL ubiquitin variant | | Descriptor: | CALCIUM ION, E3 ubiquitin-protein ligase CBL, Tyrosine protein kinase ZAP70 peptide, ... | | Authors: | Gabrielsen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2017-06-08 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | A General Strategy for Discovery of Inhibitors and Activators of RING and U-box E3 Ligases with Ubiquitin Variants.
Mol. Cell, 68, 2017
|
|
7SIY
 
 | | cCBL TKB domain in complex with pZAP70 peptide | | Descriptor: | E3 ubiquitin-protein ligase CBL, MAGNESIUM ION, Peptide from Tyrosine-protein kinase ZAP-70 | | Authors: | Murray, J.M, Yu, C. | | Deposit date: | 2021-10-15 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | cCBL TKB domain in complex with pZAP70 peptide
To Be Published
|
|
1B47
 
 | | STRUCTURE OF THE N-TERMINAL DOMAIN OF CBL IN COMPLEX WITH ITS BINDING SITE IN ZAP-70 | | Descriptor: | CALCIUM ION, CBL | | Authors: | Meng, W, Sawasdikosol, S, Burakoff, S.J, Eck, M.J. | | Deposit date: | 1999-01-06 | | Release date: | 1999-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase.
Nature, 398, 1999
|
|
8GCY
 
 | | Co-crystal structure of CBL-B in complex with N-Aryl isoindolin-1-one inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-{3-[(1s,3R)-3-methyl-1-(4-methyl-4H-1,2,4-triazol-3-yl)cyclobutyl]phenyl}-6-{[(3S)-3-methylpiperidin-1-yl]methyl}-4-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, E3 ubiquitin-protein ligase CBL-B, ... | | Authors: | Kimani, S, Zeng, H, Dong, A, Li, Y, Santhakumar, V, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The co-crystal structure of Cbl-b and a small-molecule inhibitor reveals the mechanism of Cbl-b inhibition.
Commun Biol, 6, 2023
|
|
4GPL
 
 | |
1FBV
 
 | | STRUCTURE OF A CBL-UBCH7 COMPLEX: RING DOMAIN FUNCTION IN UBIQUITIN-PROTEIN LIGASES | | Descriptor: | SIGNAL TRANSDUCTION PROTEIN CBL, SULFATE ION, UBIQUITIN-CONJUGATING ENZYME E12-18 KDA UBCH7, ... | | Authors: | Zheng, N, Wang, P, Jeffrey, P.D, Pavletich, N.P. | | Deposit date: | 2000-07-17 | | Release date: | 2000-08-30 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases.
Cell(Cambridge,Mass.), 102, 2000
|
|
6BK5
 
 | |
6O02
 
 | |
1YVH
 
 | |
6O03
 
 | |
2Y1M
 
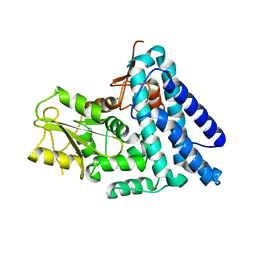 | | Structure of native c-Cbl | | Descriptor: | CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE, ZINC ION | | Authors: | Dou, H, Sibbet, G.J, Huang, D.T. | | Deposit date: | 2010-12-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural Basis for Autoinhibition and Phosphorylation-Dependent Activation of C-Cbl.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2Y1N
 
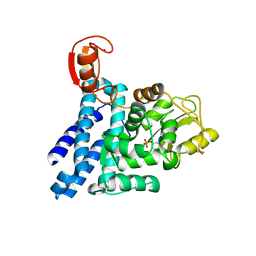 | | Structure of c-Cbl-ZAP-70 peptide complex | | Descriptor: | CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE, TYROSINE-PROTEIN KINASE ZAP-70 ZAP-70,70 KDA ZETA-ASSOCIATED PROTEIN, ... | | Authors: | Dou, H, Sibbet, G.J, Huang, D.T. | | Deposit date: | 2010-12-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural Basis for Autoinhibition and Phosphorylation-Dependent Activation of C-Cbl.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2CBL
 
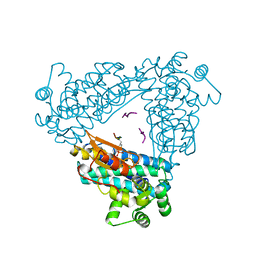 | | N-TERMINAL DOMAIN OF CBL IN COMPLEX WITH ITS BINDING SITE ON ZAP-70 | | Descriptor: | CALCIUM ION, PROTO-ONCOGENE CBL, ZAP-70 | | Authors: | Meng, W, Sawasdikosol, S, Burakoff, S.J, Eck, M.J. | | Deposit date: | 1998-08-28 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase.
Nature, 398, 1999
|
|
3ZNI
 
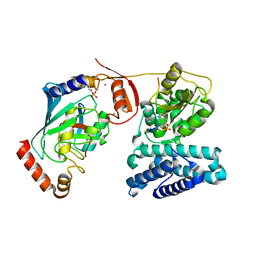 | | Structure of phosphoTyr363-Cbl-b - UbcH5B-Ub - ZAP-70 peptide complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE CBL-B, ... | | Authors: | Dou, H, Buetow, L, Sibbet, G.J, Cameron, K, Huang, D.T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Essentiality of a Non-Ring Element in Priming Donor Ubiquitin for Catalysis by a Monomeric E3.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4A4B
 
 | | Structure of modified phosphoTyr371-c-Cbl-UbcH5B-ZAP-70 complex | | Descriptor: | CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE CBL, TYROSINE-PROTEIN KINASE ZAP-70, ... | | Authors: | Dou, H, Buetow, L, Hock, A, Sibbet, G.J, Vousden, K.H, Huang, D.T. | | Deposit date: | 2011-10-08 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.789 Å) | | Cite: | Structural Basis for Autoinhibition and Phosphorylation-Dependent Activation of C-Cbl
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A4C
 
 | | Structure of phosphoTyr371-c-Cbl-UbcH5B-ZAP-70 complex | | Descriptor: | CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE CBL, TYROSINE-PROTEIN KINASE ZAP-70, ... | | Authors: | Dou, H, Buetow, L, Hock, A, Sibbet, G.J, Vousden, K.H, Huang, D.T. | | Deposit date: | 2011-10-08 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Structural Basis for Autoinhibition and Phosphorylation-Dependent Activation of C-Cbl
Nat.Struct.Mol.Biol., 19, 2012
|
|
3BUN
 
 | | Crystal structure of c-Cbl-TKB domain complexed with its binding motif in Sprouty4 | | Descriptor: | 13-meric peptide from Protein sprouty homolog 4, E3 ubiquitin-protein ligase CBL | | Authors: | Ng, C, Jackson, R.A, Buschdorf, J.P, Sun, Q, Guy, G.R, Sivaraman, J. | | Deposit date: | 2008-01-03 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates
Embo J., 27, 2008
|
|
3BUX
 
 | | Crystal structure of c-Cbl-TKB domain complexed with its binding motif in c-Met | | Descriptor: | 13-meric peptide from Hepatocyte growth factor receptor, E3 ubiquitin-protein ligase CBL | | Authors: | Ng, C, Jackson, R.A, Buschdorf, J.P, Sun, Q, Guy, G.R, Sivaraman, J. | | Deposit date: | 2008-01-03 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates
Embo J., 27, 2008
|
|
3BUM
 
 | | Crystal structure of c-Cbl-TKB domain complexed with its binding motif in Sprouty2 | | Descriptor: | E3 ubiquitin-protein ligase CBL, Protein sprouty homolog 2 | | Authors: | Ng, C, Jackson, A.R, Buschdorf, P.J, Sun, Q, Guy, R.G, Sivaraman, J. | | Deposit date: | 2008-01-03 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates
Embo J., 27, 2008
|
|
3BUW
 
 | | Crystal structure of c-Cbl-TKB domain complexed with its binding motif in Syk | | Descriptor: | 13-meric peptide from Tyrosine-protein kinase SYK, E3 ubiquitin-protein ligase CBL | | Authors: | Ng, C, Jackson, R.A, Buschdorf, J.P, Sun, Q, Guy, G.R, Sivaraman, J. | | Deposit date: | 2008-01-03 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates
Embo J., 27, 2008
|
|
3BUO
 
 | | Crystal structure of c-Cbl-TKB domain complexed with its binding motif in EGF receptor' | | Descriptor: | 13-meric peptide from Epidermal growth factor receptor, E3 ubiquitin-protein ligase CBL | | Authors: | Ng, C, Jackson, R.A, Buschdorf, J.P, Sun, Q, Guy, G.R, Sivaraman, J. | | Deposit date: | 2008-01-03 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates
Embo J., 27, 2008
|
|
3OB1
 
 | |
3OB2
 
 | |
