2MOW
 
 | |
2MOX
 
 | | solution structure of tandem SH3 domain of Sorbin and SH3 domain-containing protein 1 | | Descriptor: | Sorbin and SH3 domain-containing protein 1 | | Authors: | Zhao, D, Wang, C, Zhang, J, Wu, J, Shi, Y, Zhang, Z, Gong, Q. | | Deposit date: | 2014-05-07 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural investigation of the interaction between the tandem SH3 domains of c-Cbl-associated protein and vinculin
J.Struct.Biol., 187, 2014
|
|
2MOZ
 
 | | Structure of the Membrane Protein MerF, a Bacterial Mercury Transporter, Improved by the Inclusion of Chemical Shift Anisotropy Constraints | | Descriptor: | MerF | | Authors: | Tian, Y, Lu, G.J, Marassi, F.M, Opella, S.J, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2014-05-07 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the membrane protein MerF, a bacterial mercury transporter, improved by the inclusion of chemical shift anisotropy constraints.
J.Biomol.Nmr, 60, 2014
|
|
2MP0
 
 | | Protein Phosphorylation upon a Fleeting Encounter | | Descriptor: | Glucose-specific phosphotransferase enzyme IIA component, PHOSPHITE ION, Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Xing, Q, Yang, J, Huang, P, Zhang, W, Tang, C. | | Deposit date: | 2014-05-08 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Visualizing an ultra-weak protein-protein interaction in phosphorylation signaling.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2MP1
 
 | |
2MP2
 
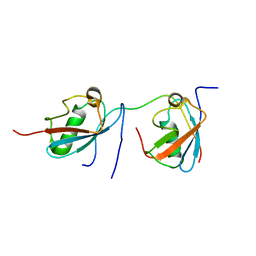 | | Solution structure of SUMO dimer in complex with SIM2-3 from RNF4 | | Descriptor: | E3 ubiquitin-protein ligase RNF4, Small ubiquitin-related modifier 3 | | Authors: | Xu, Y, Plechanovov, A, Simpson, P, Marchant, J, Leidecker, O, Sebastian, K, Hay, R.T, Matthews, S.J. | | Deposit date: | 2014-05-09 | | Release date: | 2014-07-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into SUMO chain recognition and manipulation by the ubiquitin ligase RNF4.
Nat Commun, 5, 2014
|
|
2MP3
 
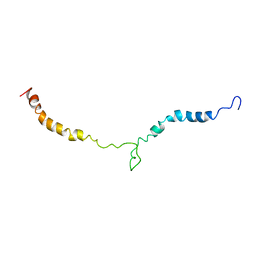 | | Truncated L126Z-sod1 in DPC micelle | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Lim, L, Song, J. | | Deposit date: | 2014-05-10 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mechanism for transforming cytosolic SOD1 into integral membrane proteins of organelles by ALS-causing mutations
Biochim.Biophys.Acta, 1848, 2015
|
|
2MP4
 
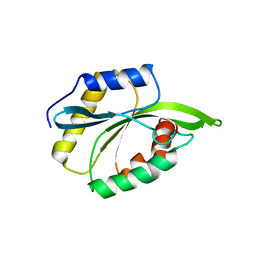 | | Solution Structure of ADF like UNC-60A Protein of Caenorhabditis elegans | | Descriptor: | Actin-depolymerizing factor 1, isoforms a/b | | Authors: | Shukla, V, Yadav, R, Kabra, A, Kumar, D, Ono, S, Arora, A. | | Deposit date: | 2014-05-11 | | Release date: | 2014-06-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Backbone dynamics of ADF like UNC-60A protein from Caenorhabditis elegans: its divergence from conventional ADF/cofilin
To be Published
|
|
2MP5
 
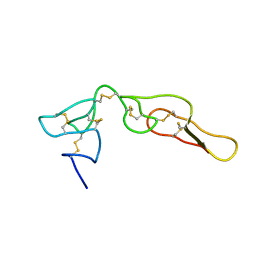 | | Structure of Bitistatin B | | Descriptor: | Disintegrin bitistatin | | Authors: | Carbajo, R.J, Calvete, J, Sanz, L, Perez, A. | | Deposit date: | 2014-05-11 | | Release date: | 2014-11-12 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of bitistatin - a missing piece in the evolutionary pathway of snake venom disintegrins.
Febs J., 282, 2015
|
|
2MP8
 
 | | NMR structure of NKR-5-3B | | Descriptor: | NKR-5-3B | | Authors: | Rosengren, K.J, Craik, D.J. | | Deposit date: | 2014-05-13 | | Release date: | 2015-05-13 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Identification, Characterization, and Three-Dimensional Structure of the Novel Circular Bacteriocin, Enterocin NKR-5-3B, from Enterococcus faecium
Biochemistry, 54, 2015
|
|
2MP9
 
 | | Solution structure of an potent antifungal peptide Cm-p5 derived from C. muricatus | | Descriptor: | Antifungal peptide | | Authors: | Sun, Z.J, Heffron, G, Mcbeth, C, Wagner, G, Otero-Gonzales, A.J, Starnbach, M.N. | | Deposit date: | 2014-05-14 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Cm-p5: an antifungal hydrophilic peptide derived from the coastal mollusk Cenchritis muricatus (Gastropoda: Littorinidae).
Faseb J., 29, 2015
|
|
2MPB
 
 | | NMR structure of BA42 protein from the psychrophilic bacteria Bizionia argentinensis sp. nov | | Descriptor: | BA42 | | Authors: | Cicero, D, Aran, M, Smal, C, Pellizza, L, Gallo, M. | | Deposit date: | 2014-05-14 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution and crystal structure of BA42, a protein from the Antarctic bacterium Bizionia argentinensis comprised of a stand-alone TPM domain.
Proteins, 82, 2014
|
|
2MPC
 
 | |
2MPE
 
 | |
2MPF
 
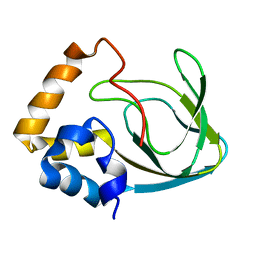 | | Solution structure human HCN2 CNBD in the cAMP-unbound state | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Saponaro, A, Pauleta, S.R, Cantini, F, Matzapetakis, M, Hammann, C, Banci, L, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2014-05-16 | | Release date: | 2014-09-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the mutual antagonism of cAMP and TRIP8b in regulating HCN channel function.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2MPG
 
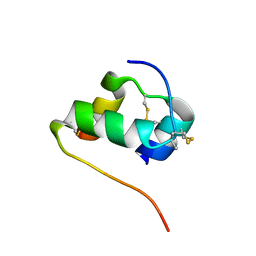 | | Solution structure of the [AibB8,LysB28,ProB29]-insulin analogue | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Kosinova, L, Jiracek, J, Zakova, L, Veverka, V. | | Deposit date: | 2014-05-17 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Insight into the structural and biological relevance of the T/R transition of the N-terminus of the B-chain in human insulin.
Biochemistry, 53, 2014
|
|
2MPH
 
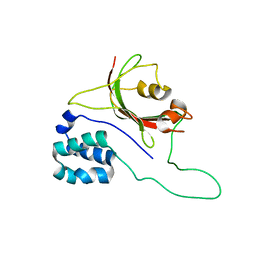 | |
2MPI
 
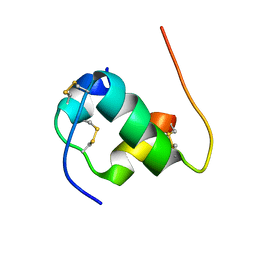 | | Solution structure of B24G insulin | | Descriptor: | insulin chain A, insulin chain B | | Authors: | Yang, Y, Wickramasinghe, N.P, Hua, Q, Weiss, M.A. | | Deposit date: | 2014-05-19 | | Release date: | 2014-12-24 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | Protective hinge in insulin opens to enable its receptor engagement.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2MPJ
 
 | | NMR structure of Xenopus RecQ4 zinc knuckle | | Descriptor: | RECQL4-helicase-like protein, ZINC ION | | Authors: | Zucchelli, C, Marino, F, Mojumdar, A, Onesti, S, Musco, G. | | Deposit date: | 2014-05-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and biochemical characterization of an RNA/DNA binding motif in the N-terminal domain of RecQ4 helicases
Sci Rep, 6, 2016
|
|
2MPK
 
 | |
2MPL
 
 | |
2MPM
 
 | | Structural Basis of Receptor Sulfotyrosine Recognition by a CC Chemokine: the N-terminal Region of CCR3 Bound to CCL11/Eotaxin-1 | | Descriptor: | CCR3, Eotaxin | | Authors: | Millard, C.J, Ludeman, J.P, Canals, M, Bridgford, J.L, Hinds, M.G, Clayton, D.J, Christopoulos, A, Payne, R.J, Stone, M.J. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-10 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Receptor Sulfotyrosine Recognition by a CC Chemokine: The N-Terminal Region of CCR3 Bound to CCL11/Eotaxin-1.
Structure, 22, 2014
|
|
2MPN
 
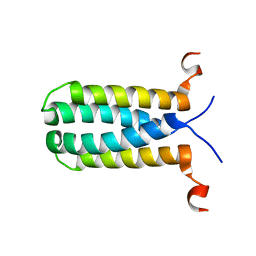 | | 3D NMR structure of the transmembrane domain of the full-length inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Bordignon, E, Maslennikov, I, Choe, S, Riek, R. | | Deposit date: | 2014-05-29 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure and Functional Analysis of the Integral Membrane Protein YgaP from Escherichia coli.
J.Biol.Chem., 289, 2014
|
|
2MPO
 
 | |
2MPQ
 
 | |
