+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2ws7 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Semi-synthetic analogue of human insulin ProB26-DTI | ||||||
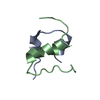
 Components Components |
| ||||||
 Keywords Keywords | HORMONE / CARBOHYDRATE METABOLISM / GLUCOSE METABOLISM / ANALOGUE / DIABETES MELLITUS | ||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of glycogen catabolic process / positive regulation of nitric oxide mediated signal transduction / negative regulation of fatty acid metabolic process / negative regulation of feeding behavior / Signaling by Insulin receptor / IRS activation / Insulin processing / regulation of protein secretion / positive regulation of peptide hormone secretion / positive regulation of respiratory burst ...negative regulation of glycogen catabolic process / positive regulation of nitric oxide mediated signal transduction / negative regulation of fatty acid metabolic process / negative regulation of feeding behavior / Signaling by Insulin receptor / IRS activation / Insulin processing / regulation of protein secretion / positive regulation of peptide hormone secretion / positive regulation of respiratory burst / Regulation of gene expression in beta cells / negative regulation of acute inflammatory response / alpha-beta T cell activation / Synthesis, secretion, and deacylation of Ghrelin / positive regulation of dendritic spine maintenance / negative regulation of protein secretion / negative regulation of gluconeogenesis / positive regulation of glycogen biosynthetic process / fatty acid homeostasis / Signal attenuation / positive regulation of insulin receptor signaling pathway / negative regulation of respiratory burst involved in inflammatory response / FOXO-mediated transcription of oxidative stress, metabolic and neuronal genes / negative regulation of lipid catabolic process / positive regulation of lipid biosynthetic process / negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway / regulation of protein localization to plasma membrane / transport vesicle / nitric oxide-cGMP-mediated signaling / COPI-mediated anterograde transport / positive regulation of nitric-oxide synthase activity / Insulin receptor recycling / negative regulation of reactive oxygen species biosynthetic process / positive regulation of brown fat cell differentiation / insulin-like growth factor receptor binding / NPAS4 regulates expression of target genes / neuron projection maintenance / endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of mitotic nuclear division / Insulin receptor signalling cascade / positive regulation of glycolytic process / positive regulation of cytokine production / endosome lumen / acute-phase response / positive regulation of long-term synaptic potentiation / positive regulation of D-glucose import across plasma membrane / positive regulation of protein secretion / insulin receptor binding / positive regulation of cell differentiation / Regulation of insulin secretion / wound healing / positive regulation of neuron projection development / hormone activity / negative regulation of protein catabolic process / regulation of synaptic plasticity / positive regulation of protein localization to nucleus / Golgi lumen / vasodilation / cognition / glucose metabolic process / insulin receptor signaling pathway / cell-cell signaling / glucose homeostasis / regulation of protein localization / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / positive regulation of cell growth / protease binding / secretory granule lumen / positive regulation of canonical NF-kappaB signal transduction / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / positive regulation of MAPK cascade / positive regulation of cell migration / G protein-coupled receptor signaling pathway / endoplasmic reticulum lumen / Amyloid fiber formation / receptor ligand activity / Golgi membrane / negative regulation of gene expression / positive regulation of cell population proliferation / positive regulation of gene expression / regulation of DNA-templated transcription / extracellular space / extracellular region / identical protein binding Similarity search - Function | ||||||
| Biological species |  HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.59 Å MOLECULAR REPLACEMENT / Resolution: 2.59 Å | ||||||
 Authors Authors | Brzozowski, A.M. / Jiracek, J. / Zakova, L. / Antolikova, E. / Watson, C.J. / Turkenburg, J.P. / Dodson, G.G. | ||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2010 Journal: Proc.Natl.Acad.Sci.USA / Year: 2010Title: Implications for the Active Form of Human Insulin Based on the Structural Convergence of Highly Active Hormone Analogues. Authors: Jiracek, J. / Zakova, L. / Antolikova, E. / Watson, C.J. / Turkenburg, J.P. / Dodson, G.G. / Brzozowski, A.M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2ws7.cif.gz 2ws7.cif.gz | 64.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2ws7.ent.gz pdb2ws7.ent.gz | 50.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2ws7.json.gz 2ws7.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ws/2ws7 https://data.pdbj.org/pub/pdb/validation_reports/ws/2ws7 ftp://data.pdbj.org/pub/pdb/validation_reports/ws/2ws7 ftp://data.pdbj.org/pub/pdb/validation_reports/ws/2ws7 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2wruC  2wrvC  2wrwC  2wrxC  2ws0C  2ws1C  2ws4C  2ws6C  1msoS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| Unit cell |
| |||||||||
| Components on special symmetry positions |
|
- Components
Components
-Protein/peptide , 2 types, 12 molecules ACEGIKBDFHJL
| #1: Protein/peptide | Mass: 2383.698 Da / Num. of mol.: 6 / Source method: obtained synthetically / Source: (synth.)  HOMO SAPIENS (human) / References: UniProt: P01308 HOMO SAPIENS (human) / References: UniProt: P01308#2: Protein/peptide | Mass: 2939.392 Da / Num. of mol.: 6 / Fragment: RESIDUES 25-50 / Mutation: YES / Source method: obtained synthetically / Source: (synth.)  HOMO SAPIENS (human) / References: UniProt: P01308 HOMO SAPIENS (human) / References: UniProt: P01308 |
|---|
-Non-polymers , 4 types, 52 molecules 






| #3: Chemical | ChemComp-IPH / #4: Chemical | #5: Chemical | #6: Water | ChemComp-HOH / | |
|---|
-Details
| Compound details | ENGINEERED RESIDUE IN CHAIN B, TYR 50 TO PRO ENGINEERED RESIDUE IN CHAIN D, TYR 50 TO PRO ...ENGINEERED |
|---|---|
| Has protein modification | Y |
| Sequence details | Y26P MUTATION 27-30 RESIDUES ARE DELETED |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.2 Å3/Da / Density % sol: 44 % / Description: NONE |
|---|---|
| Crystal grow | pH: 7.5 Details: 5 MM ZN ACETATE,35 MM NA CITRATE,0.7% PHENOL,).7M NACL,0.3M TRIS PH 7.5 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID23-2 / Wavelength: 0.8726 / Beamline: ID23-2 / Wavelength: 0.8726 |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Dec 4, 2008 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.8726 Å / Relative weight: 1 |
| Reflection | Resolution: 2.6→50 Å / Num. obs: 7871 / % possible obs: 96.6 % / Observed criterion σ(I): 0 / Redundancy: 3.3 % / Biso Wilson estimate: 31.6 Å2 / Rmerge(I) obs: 0.06 / Net I/σ(I): 14.7 |
| Reflection shell | Resolution: 2.6→2.64 Å / Redundancy: 2.5 % / Rmerge(I) obs: 0.15 / Mean I/σ(I) obs: 10.5 / % possible all: 78.1 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1MSO Resolution: 2.59→57.26 Å / Cor.coef. Fo:Fc: 0.921 / Cor.coef. Fo:Fc free: 0.812 / SU B: 26.892 / SU ML: 0.308 / TLS residual ADP flag: LIKELY RESIDUAL / Cross valid method: THROUGHOUT / ESU R: 0.842 / ESU R Free: 0.462 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS. U VALUES RESIDUAL ONLY. FOLLOWING RESIDUES ARE NOT MODELLED DUE TO DISORDER A1,B20-B26,D24-D26,E21,J22- -J26,K21,L1,L23-L26. FOLLOWING SIDE ...Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS. U VALUES RESIDUAL ONLY. FOLLOWING RESIDUES ARE NOT MODELLED DUE TO DISORDER A1,B20-B26,D24-D26,E21,J22- -J26,K21,L1,L23-L26. FOLLOWING SIDE CHAINS OCCUPANCIES ARE SET TO ZERO DUE TO HIGH MOBILITY D17,D21,F1,H21,J4,J17,K4, K5,L22.ATOM RECORD CONTAINS RESIDUAL B FACTORS ONLY.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 20.922 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.59→57.26 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller










































































 PDBj
PDBj



















