-Search query
-Search result
Showing 1 - 50 of 221 items for (author: langer & t)
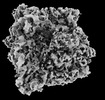
EMDB-18036: 
In situ structure of E. coli 70S ribosome
Method: subtomogram averaging / : Khusainov I, Romanov N, Goemans C, Turonova B, Zimmerli CE, Welsch S, Langer JD, Typas A, Beck M
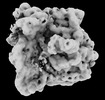
EMDB-18037: 
In situ 70S ribosome of E. coli K-12 untreated cells
Method: subtomogram averaging / : Khusainov I, Romanov N, Goemans C, Turonova B, Zimmerli CE, Welsch S, Langer JD, Typas A, Beck M

EMDB-18038: 
In situ 70S ribosome of E. coli K-12 cells treated with tetracycline
Method: subtomogram averaging / : Khusainov I, Romanov N, Goemans C, Turonova B, Zimmerli CE, Welsch S, Langer JD, Typas A, Beck M

EMDB-18039: 
In situ 70S ribosome of E. coli ED1a untreated cells
Method: subtomogram averaging / : Khusainov I, Romanov N, Goemans C, Turonova B, Zimmerli CE, Welsch S, Langer JD, Typas A, Beck M

EMDB-18040: 
In situ 70S ribosome of E. coli ED1a cells treated with tetracycline
Method: subtomogram averaging / : Khusainov I, Romanov N, Goemans C, Turonova B, Zimmerli CE, Welsch S, Langer JD, Typas A, Beck M
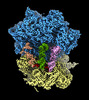
EMDB-18041: 
E. coli K-12 70S ribosome bound to mRNA A-tRNA, P-tRNA, E-tRNA
Method: single particle / : Khusainov I, Romanov N, Goemans C, Turonova B, Zimmerli CE, Welsch S, Langer JD, Typas A, Beck M
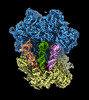
EMDB-18042: 
E. coli ED1a 70S ribosome bound to mRNA A-tRNA, P-tRNA, E-tRNA
Method: single particle / : Khusainov I, Romanov N, Goemans C, Turonova B, Zimmerli CE, Welsch S, Langer JD, Typas A, Beck M

EMDB-19206: 
E. coli ED1a 70S-tetracycline complex - focused refinement on 30S head
Method: single particle / : Khusainov I, Romanov N, Goemans C, Turonova B, Zimmerli CE, Welsch S, Langer JD, Typas A, Beck M

EMDB-19207: 
E. coli ED1a 70S-tetracycline complex - focused refinement on 30S body
Method: single particle / : Khusainov I, Romanov N, Goemans C, Turonova B, Zimmerli CE, Welsch S, Langer JD, Typas A, Beck M

EMDB-19208: 
E. coli ED1a 70S-tetracycline complex - focused refinement on 50S
Method: single particle / : Khusainov I, Romanov N, Goemans C, Turonova B, Zimmerli CE, Welsch S, Langer JD, Typas A, Beck M

EMDB-19450: 
Human UPF1 RNA helicase with AMPPNP
Method: single particle / : Langer LM, Conti E

EMDB-19451: 
Human UPF1 RNA helicase with AMPPNP and RNA
Method: single particle / : Langer LM, Conti E

EMDB-14863: 
Bacteriophage T5 head - pb10-N-Ter fused to OVA
Method: single particle / : Schoehn G, Boulanger P, Benihoud K

EMDB-16454: 
Electron cryo-tomography of HeLa cells expressing untagged Cidec
Method: electron tomography / : Ganeva I, Lim K, Boulanger J, Hoffmann PC, Muriel O, Borgeaud AC, Hagen WJH, Savage DB, Kukulski W

EMDB-16455: 
Electron cryo-tomography of HeLa cells expressing Cidec-EGFP
Method: electron tomography / : Ganeva I, Lim K, Boulanger J, Hoffmann PC, Muriel O, Borgeaud AC, Hagen WJH, Savage DB, Kukulski W

EMDB-15274: 
Single Particle cryo-EM of the lipid binding protein P116 (MPN213) from Mycoplasma pneumoniae at 3.3 Angstrom resolution.
Method: single particle / : Sprankel L, Vizarraga D

EMDB-15275: 
Single Particle cryo-EM of the empty lipid binding protein P116 (MPN213) from Mycoplasma pneumoniae at 4 Angstrom resolution
Method: single particle / : Sprankel L, Vizarraga D

EMDB-15276: 
Single Particle cryo-EM of the lipid binding protein P116 (MPN213) refilled with FBS from Mycoplasma pneumoniae at 3.5 Angstrom resolution.
Method: single particle / : Sprankel L, Vizarraga D

EMDB-15277: 
Single Particle cryo-EM of the lipid binding protein P116 (MPN213) from Mycoplasma pneumoniae bound to HDL.
Method: single particle / : Sprankel L, Vizarraga D

PDB-8a9a: 
Single Particle cryo-EM of the lipid binding protein P116 (MPN213) from Mycoplasma pneumoniae at 3.3 Angstrom resolution.
Method: single particle / : Sprankel L, Vizarraga D

PDB-8a9b: 
Single Particle cryo-EM of the empty lipid binding protein P116 (MPN213) from Mycoplasma pneumoniae at 4 Angstrom resolution
Method: single particle / : Sprankel L, Vizarraga D
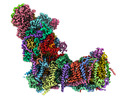
EMDB-14791: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (inhibited by DDM)
Method: single particle / : Laube E, Kuehlbrandt W

EMDB-14792: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (inhibited by DDM) - membrane arm
Method: single particle / : Laube E, Kuehlbrandt W
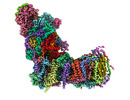
EMDB-14794: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 2)
Method: single particle / : Laube E, Kuehlbrandt W
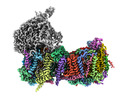
EMDB-14796: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 2) - membrane arm
Method: single particle / : Laube E, Kuehlbrandt W
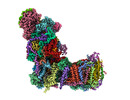
EMDB-14797: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 1)
Method: single particle / : Laube E, Kuehlbrandt W
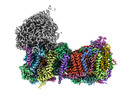
EMDB-14798: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 1) - membrane arm
Method: single particle / : Laube E, Kuehlbrandt W

PDB-7zm7: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (inhibited by DDM)
Method: single particle / : Laube E, Kuehlbrandt W

PDB-7zm8: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (inhibited by DDM) - membrane arm
Method: single particle / : Laube E, Kuehlbrandt W

PDB-7zmb: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 2)
Method: single particle / : Laube E, Kuehlbrandt W

PDB-7zme: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 2) - membrane arm
Method: single particle / : Laube E, Kuehlbrandt W

PDB-7zmg: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 1)
Method: single particle / : Laube E, Kuehlbrandt W

PDB-7zmh: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 1) - membrane arm
Method: single particle / : Laube E, Kuehlbrandt W
EMDB-13586: 
Cryo-electron tomography of ASC signalling sites in pyroptotic cells (2)
Method: electron tomography / : Liu Y, Modis Y, Alemayehu H, Hopkins LJ, Borgeaud AC, Heroven C, Howe JD, Boulanger J, Bryant CE, Zhai H

EMDB-13585: 
Cryo-electron tomography of ASC signalling sites in pyroptotic cells
Method: electron tomography / : Liu Y, Zhai H, Alemayehu H, Hopkins LJ, Borgeaud AC, Heroven C, Howe JD, Boulanger J, Bryant CE, Modis Y

EMDB-13674: 
Human SMG1-8-9 kinase complex bound to a SMG1 inhibitor
Method: single particle / : Langer LM, Conti E

EMDB-13675: 
Human SMG1-8-9 kinase complex with AlphaFold predicted SMG8 C-terminus, bound to a SMG1 inhibitor
Method: single particle / : Langer LM, Conti E

EMDB-13676: 
Human SMG1-8-9 kinase complex bound to a SMG1 inhibitor - SMG1 body
Method: single particle / : Langer LM, Conti E

EMDB-13677: 
Human SMG1-9 kinase complex bound to a SMG1 inhibitor
Method: single particle / : Langer LM, Conti E

EMDB-13678: 
Human SMG1-8-9 kinase complex bound to AMPPNP
Method: single particle / : Langer LM, Conti E

EMDB-13679: 
Human SMG1-9 kinase complex bound to AMPPNP
Method: single particle / : Langer LM, Conti E

PDB-7pw4: 
Human SMG1-8-9 kinase complex bound to a SMG1 inhibitor
Method: single particle / : Langer LM, Conti E

PDB-7pw5: 
Human SMG1-8-9 kinase complex with AlphaFold predicted SMG8 C-terminus, bound to a SMG1 inhibitor
Method: single particle / : Langer LM, Conti E

PDB-7pw6: 
Human SMG1-8-9 kinase complex bound to a SMG1 inhibitor - SMG1 body
Method: single particle / : Langer LM, Conti E

PDB-7pw7: 
Human SMG1-9 kinase complex bound to a SMG1 inhibitor
Method: single particle / : Langer LM, Conti E

PDB-7pw8: 
Human SMG1-8-9 kinase complex bound to AMPPNP
Method: single particle / : Langer LM, Conti E

EMDB-12335: 
Structure of PSII-M
Method: single particle / : Zabret J, Bohn S

EMDB-12336: 
Structure of PSII-I (PSII with Psb27, Psb28, and Psb34)
Method: single particle / : Zabret J, Bohn S

EMDB-12337: 
Structure of PSII-I prime (PSII with Psb28, and Psb34)
Method: single particle / : Zabret J, Bohn S
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
