+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ 70S ribosome of E. coli ED1a untreated cells | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cryo-ET / subtomogram averaging / ribosome / 70S / E. coli | |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 7.5 Å | |||||||||
 Authors Authors | Khusainov I / Romanov N / Goemans C / Turonova B / Zimmerli CE / Welsch S / Langer JD / Typas A / Beck M | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Bactericidal effect of tetracycline in E. coli strain ED1a may be associated with ribosome dysfunction. Authors: Iskander Khusainov / Natalie Romanov / Camille Goemans / Beata Turoňová / Christian E Zimmerli / Sonja Welsch / Julian D Langer / Athanasios Typas / Martin Beck /    Abstract: Ribosomes translate the genetic code into proteins. Recent technical advances have facilitated in situ structural analyses of ribosome functional states inside eukaryotic cells and the minimal ...Ribosomes translate the genetic code into proteins. Recent technical advances have facilitated in situ structural analyses of ribosome functional states inside eukaryotic cells and the minimal bacterium Mycoplasma. However, such analyses of Gram-negative bacteria are lacking, despite their ribosomes being major antimicrobial drug targets. Here we compare two E. coli strains, a lab E. coli K-12 and human gut isolate E. coli ED1a, for which tetracycline exhibits bacteriostatic and bactericidal action, respectively. Using our approach for close-to-native E. coli sample preparation, we assess the two strains by cryo-ET and visualize their ribosomes at high resolution in situ. Upon tetracycline treatment, these exhibit virtually identical drug binding sites, yet the conformation distribution of ribosomal complexes differs. While K-12 retains ribosomes in a translation-competent state, tRNAs are lost in the vast majority of ED1a ribosomes. These structural findings together with the proteome-wide abundance and thermal stability assessments indicate that antibiotic responses are complex in cells and can differ between different strains of a single species, thus arguing that all relevant bacterial strains should be analyzed in situ when addressing antibiotic mode of action. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18039.map.gz emd_18039.map.gz | 31.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18039-v30.xml emd-18039-v30.xml emd-18039.xml emd-18039.xml | 15 KB 15 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_18039.png emd_18039.png | 86.4 KB | ||
| Filedesc metadata |  emd-18039.cif.gz emd-18039.cif.gz | 4.4 KB | ||
| Others |  emd_18039_half_map_1.map.gz emd_18039_half_map_1.map.gz emd_18039_half_map_2.map.gz emd_18039_half_map_2.map.gz | 17.1 MB 17.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18039 http://ftp.pdbj.org/pub/emdb/structures/EMD-18039 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18039 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18039 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18039.map.gz / Format: CCP4 / Size: 33.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18039.map.gz / Format: CCP4 / Size: 33.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
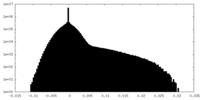
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.394 Å | ||||||||||||||||||||||||||||||||||||
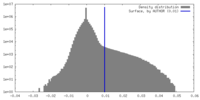
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_18039_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
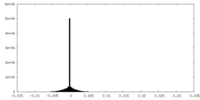
| Density Histograms |
-Half map: #2
| File | emd_18039_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
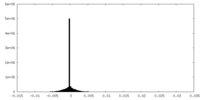
| Density Histograms |
- Sample components
Sample components
-Entire : In situ structure of 70S ribosome of untreated E. coli ED1a cells...
| Entire | Name: In situ structure of 70S ribosome of untreated E. coli ED1a cells determined by subtomogram averaging |
|---|---|
| Components |
|
-Supramolecule #1: In situ structure of 70S ribosome of untreated E. coli ED1a cells...
| Supramolecule | Name: In situ structure of 70S ribosome of untreated E. coli ED1a cells determined by subtomogram averaging type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.5 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 6.5 / Details: LB medium |
|---|---|
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 3.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 53000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 7.5 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: Warp (ver. 1.0.9) / Number subtomograms used: 30548 |
|---|---|
| Extraction | Number tomograms: 26 / Number images used: 52000 / Reference model: 70S average derived from manual picking / Method: template matching / Software - Name: Warp (ver. 1.0.9) |
| Final 3D classification | Software - Name: RELION (ver. 3.1.3) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 3.1.3) |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)