+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7pw7 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human SMG1-9 kinase complex bound to a SMG1 inhibitor | |||||||||||||||
 Components Components |
| |||||||||||||||
 Keywords Keywords | TRANSFERASE / Complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationdiacylglycerol-dependent serine/threonine kinase activity / chromatoid body / eye development / nuclear-transcribed mRNA catabolic process, nonsense-mediated decay / regulation of telomere maintenance / telomeric DNA binding / phosphatidylinositol phosphate biosynthetic process / mRNA export from nucleus / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / peptidyl-serine phosphorylation ...diacylglycerol-dependent serine/threonine kinase activity / chromatoid body / eye development / nuclear-transcribed mRNA catabolic process, nonsense-mediated decay / regulation of telomere maintenance / telomeric DNA binding / phosphatidylinositol phosphate biosynthetic process / mRNA export from nucleus / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / peptidyl-serine phosphorylation / brain development / protein autophosphorylation / heart development / in utero embryonic development / protein kinase activity / non-specific serine/threonine protein kinase / protein serine kinase activity / DNA repair / protein serine/threonine kinase activity / DNA damage response / negative regulation of apoptotic process / RNA binding / nucleoplasm / ATP binding / metal ion binding / identical protein binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.59 Å | |||||||||||||||
 Authors Authors | Langer, L.M. / Conti, E. | |||||||||||||||
| Funding support |  Germany, 1items Germany, 1items
| |||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: Cryo-EM reconstructions of inhibitor-bound SMG1 kinase reveal an autoinhibitory state dependent on SMG8. Authors: Lukas M Langer / Fabien Bonneau / Yair Gat / Elena Conti /  Abstract: The PI3K-related kinase (PIKK) SMG1 monitors the progression of metazoan nonsense-mediated mRNA decay (NMD) by phosphorylating the RNA helicase UPF1. Previous work has shown that the activity of SMG1 ...The PI3K-related kinase (PIKK) SMG1 monitors the progression of metazoan nonsense-mediated mRNA decay (NMD) by phosphorylating the RNA helicase UPF1. Previous work has shown that the activity of SMG1 is impaired by small molecule inhibitors, is reduced by the SMG1 interactors SMG8 and SMG9, and is downregulated by the so-called SMG1 insertion domain. However, the molecular basis for this complex regulatory network has remained elusive. Here, we present cryo-electron microscopy reconstructions of human SMG1-9 and SMG1-8-9 complexes bound to either a SMG1 inhibitor or a non-hydrolyzable ATP analog at overall resolutions ranging from 2.8 to 3.6 Å. These structures reveal the basis with which a small molecule inhibitor preferentially targets SMG1 over other PIKKs. By comparison with our previously reported substrate-bound structure (Langer et al.,2020), we show that the SMG1 insertion domain can exert an autoinhibitory function by directly blocking the substrate-binding path as well as overall access to the SMG1 kinase active site. Together with biochemical analysis, our data indicate that SMG1 autoinhibition is stabilized by the presence of SMG8. Our results explain the specific inhibition of SMG1 by an ATP-competitive small molecule, provide insights into regulation of its kinase activity within the NMD pathway, and expand the understanding of PIKK regulatory mechanisms in general. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7pw7.cif.gz 7pw7.cif.gz | 423 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7pw7.ent.gz pdb7pw7.ent.gz | 305.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7pw7.json.gz 7pw7.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pw/7pw7 https://data.pdbj.org/pub/pdb/validation_reports/pw/7pw7 ftp://data.pdbj.org/pub/pdb/validation_reports/pw/7pw7 ftp://data.pdbj.org/pub/pdb/validation_reports/pw/7pw7 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  13677MC  7pw4C  7pw5C  7pw6C  7pw8C  7pw9C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 2 types, 2 molecules AC
| #1: Protein | Mass: 397395.375 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: SMG1, ATX, KIAA0421, LIP / Production host: Homo sapiens (human) / Gene: SMG1, ATX, KIAA0421, LIP / Production host:  Homo sapiens (human) Homo sapiens (human)References: UniProt: Q96Q15, non-specific serine/threonine protein kinase |
|---|---|
| #2: Protein | Mass: 57717.473 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: SMG9, C19orf61 / Production host: Homo sapiens (human) / Gene: SMG9, C19orf61 / Production host:  Homo sapiens (human) / References: UniProt: Q9H0W8 Homo sapiens (human) / References: UniProt: Q9H0W8 |
-Non-polymers , 4 types, 4 molecules 
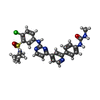





| #3: Chemical | ChemComp-IHP / |
|---|---|
| #4: Chemical | ChemComp-88C / |
| #5: Chemical | ChemComp-ATP / |
| #6: Chemical | ChemComp-MG / |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | N |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: SMG1-SMG9 kinase complex bound to a SMG1 inhibitor / Type: COMPLEX / Entity ID: #1-#2 / Source: RECOMBINANT |
|---|---|
| Molecular weight | Value: 0.597 MDa / Experimental value: NO |
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Source (recombinant) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Buffer solution | pH: 7.4 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD |
| Image recording | Electron dose: 89.32 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.19.2_4158: / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software | Name: PHENIX / Category: model refinement | ||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.59 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 85216 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller
















 PDBj
PDBj












