+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4264 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM Structure of INO80core | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | DNA BINDING PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationDASH complex / protein transport along microtubule to mitotic spindle pole body / mitotic sister chromatid biorientation / attachment of spindle microtubules to kinetochore / attachment of mitotic spindle microtubules to kinetochore / Swr1 complex / Ino80 complex / ATP-dependent activity, acting on DNA / helicase activity / kinetochore ...DASH complex / protein transport along microtubule to mitotic spindle pole body / mitotic sister chromatid biorientation / attachment of spindle microtubules to kinetochore / attachment of mitotic spindle microtubules to kinetochore / Swr1 complex / Ino80 complex / ATP-dependent activity, acting on DNA / helicase activity / kinetochore / mitotic spindle / chromatin organization / DNA helicase / chromatin remodeling / DNA repair / ATP hydrolysis activity / ATP binding Similarity search - Function | ||||||||||||
| Biological species |  Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) | ||||||||||||
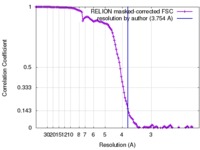
| Method | single particle reconstruction / cryo EM / Resolution: 3.754 Å | ||||||||||||
 Authors Authors | Eustermann S / Schall K | ||||||||||||
| Funding support |  Germany, 3 items Germany, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2018 Journal: Nature / Year: 2018Title: Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Authors: Sebastian Eustermann / Kevin Schall / Dirk Kostrewa / Kristina Lakomek / Mike Strauss / Manuela Moldt / Karl-Peter Hopfner /  Abstract: In the eukaryotic nucleus, DNA is packaged in the form of nucleosomes, each of which comprises about 147 base pairs of DNA wrapped around a histone protein octamer. The position and histone ...In the eukaryotic nucleus, DNA is packaged in the form of nucleosomes, each of which comprises about 147 base pairs of DNA wrapped around a histone protein octamer. The position and histone composition of nucleosomes is governed by ATP-dependent chromatin remodellers such as the 15-subunit INO80 complex . INO80 regulates gene expression, DNA repair and replication by sliding nucleosomes, the exchange of histone H2A.Z with H2A, and the positioning of + 1 and -1 nucleosomes at promoter DNA. The structures and mechanisms of these remodelling reactions are currently unknown. Here we report the cryo-electron microscopy structure of the evolutionarily conserved core of the INO80 complex from the fungus Chaetomium thermophilum bound to a nucleosome, at a global resolution of 4.3 Å and with major parts at 3.7 Å. The INO80 core cradles one entire gyre of the nucleosome through multivalent DNA and histone contacts. An Rvb1/Rvb2 AAA ATPase heterohexamer is an assembly scaffold for the complex and acts as a 'stator' for the motor and nucleosome-gripping subunits. The Swi2/Snf2 ATPase motor binds to nucleosomal DNA at superhelical location -6, unwraps approximately 15 base pairs, disrupts the H2A-DNA contacts and is poised to pump entry DNA into the nucleosome. Arp5 and Ies6 bind superhelical locations -2 and -3 to act as a counter grip for the motor, on the other side of the H2A-H2B dimer. The Arp5 insertion domain forms a grappler element that binds the nucleosome dyad, connects the Arp5 actin-fold and entry DNA over a distance of about 90 Å and packs against histone H2A-H2B near the 'acidic patch'. Our structure together with biochemical data suggests a unified mechanism for nucleosome sliding and histone editing by INO80. The motor is part of a macromolecular ratchet, persistently pumping entry DNA across the H2A-H2B dimer against the Arp5 grip until a large nucleosome translocation step occurs. The transient exposure of H2A-H2B by motor activity as well as differential recognition of H2A.Z and H2A may regulate histone exchange. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4264.map.gz emd_4264.map.gz | 88.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4264-v30.xml emd-4264-v30.xml emd-4264.xml emd-4264.xml | 23.5 KB 23.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4264_fsc.xml emd_4264_fsc.xml | 12.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_4264.png emd_4264.png | 48.6 KB | ||
| Filedesc metadata |  emd-4264.cif.gz emd-4264.cif.gz | 8.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4264 http://ftp.pdbj.org/pub/emdb/structures/EMD-4264 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4264 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4264 | HTTPS FTP |
-Related structure data
| Related structure data |  6fhsMC  4277C  4278C  4280C  6fmlC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4264.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4264.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : INO80core masked reconstruction
| Entire | Name: INO80core masked reconstruction |
|---|---|
| Components |
|
-Supramolecule #1: INO80core masked reconstruction
| Supramolecule | Name: INO80core masked reconstruction / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 Details: 3xRvb1, 3x Rvb2, Ino80insert, Arp5, Ies2, Ies6 other parts of complex are masked out |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) |
| Molecular weight | Theoretical: 600 KDa |
-Macromolecule #1: RuvB-like helicase
| Macromolecule | Name: RuvB-like helicase / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) Chaetomium thermophilum var. thermophilum DSM 1495 (fungus)Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 50.451848 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MVQISEVRGN TRDHRTAAHT HIKGLGLNSS GIAEKQAAGF VGQCAAREAC GVVVDLIKAH KMAGRGVLLA GGPGTGKTAL ALAISQELG TKIPFCPITG SEIYSTEVKK TEVLMENFRR AIGLRVRETK DVYEGEVTEM TPEEAENPLG GYGKTISTLL I GLKSARGQ ...String: MVQISEVRGN TRDHRTAAHT HIKGLGLNSS GIAEKQAAGF VGQCAAREAC GVVVDLIKAH KMAGRGVLLA GGPGTGKTAL ALAISQELG TKIPFCPITG SEIYSTEVKK TEVLMENFRR AIGLRVRETK DVYEGEVTEM TPEEAENPLG GYGKTISTLL I GLKSARGQ KKLRLDPSIY EAIQKERVQV GDVIYIETNT GACKRVGRSD AYATEFDLEA EEYVPIPKGE VHKKKEIVQD VT LHDLDVA NARPQGGQDI ISMMGQLMKP KMTEITDKLR MEINKVVQKY INQGVAELIP GVLFIDEAHM LDIECFTYLN KAL ESPIAP IVVLASNRGI ATIRGADDLK AAHGIPPDFL QRLLIIPTHP YEPDEIRRIV RIRAQTEGVQ LTDAAVDRVA EHGV RISLR YCLQLLAPAS ILARVNGRTQ VDVQDIAEAE ELFLDARRSA NILTSTGESG GLHGFIS UniProtKB: RuvB-like helicase |
-Macromolecule #2: RuvB-like helicase
| Macromolecule | Name: RuvB-like helicase / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) Chaetomium thermophilum var. thermophilum DSM 1495 (fungus)Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 53.212746 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAAPLVTSVT ETKELRGLNL IAAHSHIRGL GVDADTLEPR PSSQGLVGQE KARKAAAVVL EMIKQGKIAG RAVLIAGPPS TGKTAIAMG MAQSLGQDVP FTTLAASEIF SLEMSKTEAL TQAFRKSIGV RIKEESEIME GEVVEIQIDR SVTGGAKQGK L TIKTTDME ...String: MAAPLVTSVT ETKELRGLNL IAAHSHIRGL GVDADTLEPR PSSQGLVGQE KARKAAAVVL EMIKQGKIAG RAVLIAGPPS TGKTAIAMG MAQSLGQDVP FTTLAASEIF SLEMSKTEAL TQAFRKSIGV RIKEESEIME GEVVEIQIDR SVTGGAKQGK L TIKTTDME AIYDMGSKMI DAMTKERVMA GDIISIDKSS GKITKLGRSY ARSRDYDAMG VDTKFLQCPE GELQKRKEVV HT VSLHEID VINSRTQGFL ALFSGDTGEI RSEIRDQINT KVAEWKEEGK AEIVPGVLFI DEVHMLDIEC FSYINRALES DLA PIVIMA SNRGVSRIRG TDYKSPHGLP LDFLDRVVII NTHPYTPDEL RQILSIRAQE EEVDLTPDAL ALLTKIGQEA GLRY ASNLI TTSQLIAAKR RAKQVGVEDV QRSFKLFYDP ARSVRFVQES EKRLIGNDGV VDFSYQGAAE AAAPTLPAAA PVDPV GGEK MDMS UniProtKB: RuvB-like helicase |
-Macromolecule #3: Ino80
| Macromolecule | Name: Ino80 / type: protein_or_peptide / ID: 3 / Details: C-terminal double FlagTag / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) |
| Molecular weight | Theoretical: 127.529781 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MTDSYATKAS NLKKTAILAS KEAKRWQLRT NKGTKDLQAR AKRVMRDMMG FWKRNEREER DLRKAAERLE LENARKEEAD REAARQRRK LNFLISQTEL YSHFISKKIK THEVERSTDH PDVATDEKDK IPEPTLNINV PEPTGPIAPK VTDFNSLDFD N EDESALQA ...String: MTDSYATKAS NLKKTAILAS KEAKRWQLRT NKGTKDLQAR AKRVMRDMMG FWKRNEREER DLRKAAERLE LENARKEEAD REAARQRRK LNFLISQTEL YSHFISKKIK THEVERSTDH PDVATDEKDK IPEPTLNINV PEPTGPIAPK VTDFNSLDFD N EDESALQA AAMANAQNAI AEAQKKAREF NKDETKLDED GEMNFQHPEL TEFEVAQPKL LNCQLKEYQL KGLNWLVNLY EQ GINGILA DEMGLGKTVQ SISVMAYLAE RYDIWGPFLV VAPASTLHNW QQEVSKFVPD FKVLPYWGTA ADRKVLRKFW DRK HTTYKK DSPFHVMITS YQLVVSDVAY FQKMKWQYMI LDEAQAIKSS QSSRWKCLLG FHCRNRLLLT GTPIQNNMQE LWAL LHFIM PSLFDSHDEF SEWFSKDIES HAQSNTKLNE DQLKRLHMIL KPFMLRRVKK HVQKELGDKI EIDVFCELSY RQRAM YQSL RNQISIMDLI EKATVGDNED SATLMNLVMQ FRKVCNHPDL FERADTSSPF FCGHFAETGS FLREGTNVAL GYSTRS LVE YRLPRLIWCD GGRLDKPGPG NLVAGFRSKY LNHMMNIWTP ENIRSSLEGI ENFTWLRFVD TSLQEAYRAS HTDVFAR AV DLASKQNRLG HMQIVYDEPE DKKWTPVHAL FQICERENPK AVAEITTEGV LRDLMNIARV KYRELGLCRL EKAARPRA S APPIEVVCDS RSAVIERENI MFHPAMRKAL FGPTPSEIKE ASFGPRPVTL YPPRALLPAP DHDKQRFTNI TVPSMARFV TDSGKLAKLD ELLRELKEGG HRVLLYFQMT RMIDLMEEYL TYRNYKYCRL DGSTKLEDRR DTVADFQTRP EIFIFLLSTR AGGLGINLT TADTVIFYDS DWNPTIDSQA MDRAHRLGQT KQVTVYRLIT RGTIEERIRK RALQKEEVQR VVITGTGSVD F SGRRPPEN RNRDIAMWLA DDEQAEMIER REKELIESGE YDKIMQQRRK GGKRKRGAAN GDTVPSLEDM YHEGEGHFDD NK GSGAATP VDADSLGRGG KRKKAGGSKK AKTTKQRLAI ADGEIDDGEI DIDYKDDDDK GTDYKDDDDK |
-Macromolecule #4: les2
| Macromolecule | Name: les2 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) Chaetomium thermophilum var. thermophilum DSM 1495 (fungus)Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 53.258902 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSTRPRRHAA QRASQAITDL ADRDRESDHS HGPISSRMSS FNSSSRSRLP GKGIASVSRS EAGGASDPEH IHLTVKLPSS KLRQATSSS GIKKAGSVGS SSSSSGGGKA AVKRARGGKR SRVLESSEEE EEENEVEVLG DEDEEEEEEE DEIEVREGEG Y DEDEEDVE ...String: MSTRPRRHAA QRASQAITDL ADRDRESDHS HGPISSRMSS FNSSSRSRLP GKGIASVSRS EAGGASDPEH IHLTVKLPSS KLRQATSSS GIKKAGSVGS SSSSSGGGKA AVKRARGGKR SRVLESSEEE EEENEVEVLG DEDEEEEEEE DEIEVREGEG Y DEDEEDVE DEDEEMQDLG EEDADGEDDE MDVDAEGEED ADGDVNMDAG VVGARATTVR AVPPAIKVTK PPKESPSNGK AA TASKAND NAVPVKRPAP DSDDESLSSL ESEPEEEVNV AGGEDAEGED DDAEGEVDAE GEEEEEEEEI EVADEDAEGE DVE QDEDED EEEEDDDDEM ISRAQTPDMS RLTARQRARL GEASGEYLKL SDEVQSKKHF TAEELSMRRA EMARRRRNLS EKRN EEIKM ETVNKLLKKQ APRTTRRAAQ AAAAAEEAEE AAKQPKRPDP MMIRWVNNKM GSVVAVPEEL LGTHAGVVFG AGPGK GLPA GKMVEEV UniProtKB: INO80 complex subunit B-like conserved region domain-containing protein |
-Macromolecule #5: les6
| Macromolecule | Name: les6 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) Chaetomium thermophilum var. thermophilum DSM 1495 (fungus)Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 23.127523 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSNPDAQSAQ AAHQALVEQL DLHSIHKTFR NPNWRPNQRR NKTIKAILGE SQRKEASSTS AVATPRADDN GGGSGADTPA NNDNNDGLS TSGTSTPANG NGSGAGTPAS NGQPNLAQAS RSLQKLVLEK SLASAQAPDK KAANGFASSA PTATYTNIES A PSLAPMKH ...String: MSNPDAQSAQ AAHQALVEQL DLHSIHKTFR NPNWRPNQRR NKTIKAILGE SQRKEASSTS AVATPRADDN GGGSGADTPA NNDNNDGLS TSGTSTPANG NGSGAGTPAS NGQPNLAQAS RSLQKLVLEK SLASAQAPDK KAANGFASSA PTATYTNIES A PSLAPMKH YCDVTGLPAP YLDPKTRLRY HNKEIFAMIR NLPQGMGEQF LEARGAHTVL K UniProtKB: Vps72/YL1 C-terminal domain-containing protein |
-Macromolecule #6: Arp5
| Macromolecule | Name: Arp5 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) Chaetomium thermophilum var. thermophilum DSM 1495 (fungus)Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 87.773086 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAPSAVAEPP PIPQRDEPWK RLPPPTVYPV KEARFEKYIP PQLDGRERAL AQPPGQVAIV IDNGSHSVRA GWNFEDKPRL AIPPIMSKY RDRKMGKTFS FAGSDCYADT TARSHIRNAF EAGTGIVSNW DVMEHVLDYV FVKLGMNECD GAIDMPIVMT E AVANLPYS ...String: MAPSAVAEPP PIPQRDEPWK RLPPPTVYPV KEARFEKYIP PQLDGRERAL AQPPGQVAIV IDNGSHSVRA GWNFEDKPRL AIPPIMSKY RDRKMGKTFS FAGSDCYADT TARSHIRNAF EAGTGIVSNW DVMEHVLDYV FVKLGMNECD GAIDMPIVMT E AVANLPYS RKSMSEIIFE CYGAPSLVYG IDSLFSFRHN QGQTGLVVSS SYSATHVIPV YNRKALLSQA IRLNWGGWHM AE YMLKLLK LKYYTGFPGK LNSSQTEHMV RDFCYVSLDY DRELAGYLDW TGLEDRERIV QYPYTEEVVV QKTEEELARI AER KKESGR RLQEQAAKMR LERLMKKEQE LEYYKDIQRR MQGESKKEIK RLLDEAELKD EAALERVIRD LERSIKRARQ KDLG EPEEE EVPDFSLLDV PDDQLDEAGL RQKRQQRLLK SNWEARQRAK AEKEAEKARL AEEARLDEER RKNDLEGWLE EKRQL RLAK LNQLKERERL KADLGNRKSL ASQIRMKNIA NLASDNPTGS GSRKRRRGGA GADQDDDFGA DDADWGVYRS VAIGAN KGD DSDDEEGEED LEAAIRSLEN DLLRYDKTFS YDMTLDAQRD WSKSLLHAFR YGPRPFDPSS QAETHRVHLN VERIRVP EV LFQPAAIAGV DQAGLVEIAG DILCQRLPSL PGIQDAPDAF LRDVFLTGGN TLFQNFDERL RQGLMALLPV GAPLRVRR A QDAILDAWRG AAGWACTEEA KAAWITREEY LEKGGEYIKE HDLGNAFA UniProtKB: Uncharacterized protein |
-Macromolecule #7: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 7 / Number of copies: 6 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #8: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 8 / Number of copies: 1 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 20 mM HEPES pH 8, 60 mM KCl, 0.5% glycerol, 0.25 mM CaCl2, 20 uM ZnCl2, 0.25 mM DTT, 0.05% Octyl-beta-glucoside |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 281 K / Instrument: LEICA EM GP |
| Details | Monodisperse sample: INO80core complex reconstituted with nucleosomal substrate was purified by gelfiltration. Addition of nucleotides or crosslinking was not required. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 3992 / Average electron dose: 59.6 e/Å2 Details: Images were collected in movie mode with 4 frames per second and 10s total aquisition |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus min: 1.3 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)