[English] 日本語
 Yorodumi
Yorodumi- EMDB-13676: Human SMG1-8-9 kinase complex bound to a SMG1 inhibitor - SMG1 body -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13676 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human SMG1-8-9 kinase complex bound to a SMG1 inhibitor - SMG1 body | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationdiacylglycerol-dependent serine/threonine kinase activity / chromatoid body / nuclear-transcribed mRNA catabolic process, nonsense-mediated decay / regulation of telomere maintenance / telomeric DNA binding / phosphatidylinositol phosphate biosynthetic process / mRNA export from nucleus / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / peptidyl-serine phosphorylation / protein autophosphorylation ...diacylglycerol-dependent serine/threonine kinase activity / chromatoid body / nuclear-transcribed mRNA catabolic process, nonsense-mediated decay / regulation of telomere maintenance / telomeric DNA binding / phosphatidylinositol phosphate biosynthetic process / mRNA export from nucleus / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / peptidyl-serine phosphorylation / protein autophosphorylation / protein kinase activity / non-specific serine/threonine protein kinase / protein serine kinase activity / DNA repair / protein serine/threonine kinase activity / DNA damage response / RNA binding / nucleoplasm / ATP binding / metal ion binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.05 Å | |||||||||
 Authors Authors | Langer LM / Conti E | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: Cryo-EM reconstructions of inhibitor-bound SMG1 kinase reveal an autoinhibitory state dependent on SMG8. Authors: Lukas M Langer / Fabien Bonneau / Yair Gat / Elena Conti /  Abstract: The PI3K-related kinase (PIKK) SMG1 monitors the progression of metazoan nonsense-mediated mRNA decay (NMD) by phosphorylating the RNA helicase UPF1. Previous work has shown that the activity of SMG1 ...The PI3K-related kinase (PIKK) SMG1 monitors the progression of metazoan nonsense-mediated mRNA decay (NMD) by phosphorylating the RNA helicase UPF1. Previous work has shown that the activity of SMG1 is impaired by small molecule inhibitors, is reduced by the SMG1 interactors SMG8 and SMG9, and is downregulated by the so-called SMG1 insertion domain. However, the molecular basis for this complex regulatory network has remained elusive. Here, we present cryo-electron microscopy reconstructions of human SMG1-9 and SMG1-8-9 complexes bound to either a SMG1 inhibitor or a non-hydrolyzable ATP analog at overall resolutions ranging from 2.8 to 3.6 Å. These structures reveal the basis with which a small molecule inhibitor preferentially targets SMG1 over other PIKKs. By comparison with our previously reported substrate-bound structure (Langer et al.,2020), we show that the SMG1 insertion domain can exert an autoinhibitory function by directly blocking the substrate-binding path as well as overall access to the SMG1 kinase active site. Together with biochemical analysis, our data indicate that SMG1 autoinhibition is stabilized by the presence of SMG8. Our results explain the specific inhibition of SMG1 by an ATP-competitive small molecule, provide insights into regulation of its kinase activity within the NMD pathway, and expand the understanding of PIKK regulatory mechanisms in general. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13676.map.gz emd_13676.map.gz | 3.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13676-v30.xml emd-13676-v30.xml emd-13676.xml emd-13676.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13676.png emd_13676.png | 49.7 KB | ||
| Filedesc metadata |  emd-13676.cif.gz emd-13676.cif.gz | 7.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13676 http://ftp.pdbj.org/pub/emdb/structures/EMD-13676 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13676 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13676 | HTTPS FTP |
-Related structure data
| Related structure data |  7pw6MC  7pw4C  7pw5C  7pw7C  7pw8C  7pw9C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13676.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13676.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8512 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : SMG1-SMG8-SMG9 kinase complex bound to a SMG1 inhibitor
| Entire | Name: SMG1-SMG8-SMG9 kinase complex bound to a SMG1 inhibitor |
|---|---|
| Components |
|
-Supramolecule #1: SMG1-SMG8-SMG9 kinase complex bound to a SMG1 inhibitor
| Supramolecule | Name: SMG1-SMG8-SMG9 kinase complex bound to a SMG1 inhibitor type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 597 KDa |
-Macromolecule #1: Serine/threonine-protein kinase SMG1,Serine/threonine-protein kin...
| Macromolecule | Name: Serine/threonine-protein kinase SMG1,Serine/threonine-protein kinase SMG1,Serine/threonine-protein kinase SMG1,Serine/threonine-protein kinase SMG1,Serine/threonine-protein kinase ...Name: Serine/threonine-protein kinase SMG1,Serine/threonine-protein kinase SMG1,Serine/threonine-protein kinase SMG1,Serine/threonine-protein kinase SMG1,Serine/threonine-protein kinase SMG1,Serine/threonine-protein kinase SMG1,Serine/threonine-protein kinase SMG1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 320.901969 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ANTLVEDVNI CLQACSSLHA LSSSLPDDLL QRCVDVCRVQ LVHSGTRIRQ AFGKLLKSIP LDVVLSNNNH TEIQEISLAL RSHMSKAPS NTFHPQDFSD VISFILYGNS HRTGKDNWLE RLFYSCQRLD KRDQSTIPRN LLKTDAVLWQ WAIWEAAQFT V LSKLRTPL ...String: ANTLVEDVNI CLQACSSLHA LSSSLPDDLL QRCVDVCRVQ LVHSGTRIRQ AFGKLLKSIP LDVVLSNNNH TEIQEISLAL RSHMSKAPS NTFHPQDFSD VISFILYGNS HRTGKDNWLE RLFYSCQRLD KRDQSTIPRN LLKTDAVLWQ WAIWEAAQFT V LSKLRTPL GRAQDTFQTI EGIIRSLAAH TLNPDQDVSQ WTTADNDEGH GNNQLRLVLL LQYLENLEKL MYNAYEGCAN AL TSPPKVI RTFFYTNRQT CQDWLTRIRL SIMRVGLLAG QPAVTVRHGF DLLTEMKTTS LSQGNELEVT IMMVVEALCE LHC PEAIQG IAVWSSSIVG KNLLWINSVA QQAEGRFEKA SVEYQEHLCA MTGVDCCISS FDKSVLTLAN AGRNSASPKH SLNG ESRKT VLSKPTDSSP EVINYLGNKA CECYISIADW AAVQEWQNSI HDLKKSTSST SLNLKADFNY IKSLSSFESG KFVEC TEQL ELLPGENINL LAGGSKEKIN MKKLLPNMLS PDPRELQKSI EVQLLRSSVC LATALNPIEQ DQKWQSITEN VVKYLK QTS RIAIGPLRLS TLTVSQSLPV LSTLQLYCSS ALENTVSNRL STEDCLIPLF SEALRSCKQH DVRPWMQALR YTMYQNQ LL EKIKEQTVPI RSHLMELGLT AAKFARKRGN VSLATRLLAQ CSEVQLGKTT TAQDLVQHFK KLSTQGQVDE KWGPELDI E KTKLLYTAGQ STHAMEMLSS CAISFCKSVK AEYAVAKSIL TLAKWIQAEW KEISGQLKQV YRAQHQQNFT GLSTLSKNI LTLIELPSVN TMEEEYPRIE SESTVHIGVG EPDFILGQLY HLSSVQAPEV AKSWAALASW AYRWGRKVVD NAS(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)EGV IKVWRKVVDR IFSLYKLSCS AYFTFLKLNA GQIPLDEDDP RL HLSHRVE QSTDDMIVMA TLRLLRLLVK HAGELRQYLE HGLETTPTAP WRGIIPQLFS RLNHPEVYVR QSICNLLCRV AQD SPHLIL YPAIVGTISL SSESQASGNK FSTAIPTLLG NIQGEELLVS ECEGGSPPAS QDSNKDEPKS GLNEDQAMMQ DCYS KIVDK LSSANPTMVL QVQMLVAELR RVTVLWDELW LGVLLQQHMY VL(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)PHEKWFQD NYGDAIENAL EKLK(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)YIL RLEEISPWLA AMTNTEIALP GEVSARDTVT IHSVGGTITI LPTKTKP KK LLFLGSDGKS YPYLFKGLED LHLDERIMQF LSIVNTMFAT INRQETPRFH ARHYSVTPLG TRSGLIQWVD GATPLFGL Y KRWQQREAAL QAQKAQDSYQ TPQNPGIVPR PSELYYSKIG PALKTVGLSL DVSRRDWPLH VMKAVLEELM EATPPNLLA KELWSSCTTP DEWWRVTQSY ARSTAVMSMV GYIIGLGDRH LDNVLIDMTT GEVVHIDYNV CFEKGKSLRV PEKVPFRMTQ NIETALGVT GVEGVFRLSC EQVLHIMRRG RETLLTLLEA FVYDPLVDWT AGGEAGFAGA VYGGGGQQAE SKQSKREMER E ITRSLFSS RVAEIKVNWF KNRDEMLVVL PKLDGSLDEY LSLQEQLTDV EKLQGKLLEE IEFLEGAEGV DHPSHTLQHR YS EHTQLQT QQRAVQEAIQ VKLNEFEQWI THYQAAFNNL EATQLASLLQ EISTQMDLGP PSYVPATAFL QNAGQAHLIS QCE QLEGEV GALLQQRRSV LRGCLEQLHH YATVALQYPK AIFQKHRIEQ WKTWMEELIC NTTVERCQEL YRKYEMQYAP QPPP TVCQF ITATEMTLQR YAADINSRLI RQVERLKQEA VTVPVCEDQL KEIERCIKVF LHENGEEGSL SLASVIISAL CTLTR RNLM MEGAASSAGE QLVDLTSRDG AWFLEELCSM SGNVTCLVQL LKQCHLVPQD LDIPNPMEAS ETVHLANGVY TSLQEL NSN FRQIIFPEAL RCLMKGEYTL ESMLHELDGL IEQTTDGVPL QTLVESLQAY LRNAAMGLEE ETHAHYIDVA RLLHAQY GE LIQPRNGSVD ETPKMSAGQM LLVAFDGMFA QVETAFSLLV EKLNKMEIPI AWRKIDIIRE ARSTQVNFFD DDNHRQVL E EIFFLKRLQT IKEFFRLCGT FSKTLSGSSS LEDQNTVNGP VQIVNVKTLF RNSCFSEDQM AKPIKAFTAD FVRQLLIGL PNQALGLTLC SFISALGVDI IAQVEAKDFG AESKVSVDDL CKKAVEHNIQ IGKFSQLVMN RATVLASSYD TAWKKHDLVR RLETSISSC KTSLQRVQLH IAMFQWQHED LLINRPQAMS VTPPPRSAIL TSMKKKLHTL SQIETSIATV QEKLAALESS I EQRLKWAG GANPALAPVL QDFEATIAER RNLVLKESQR ASQVTFLCSN IIHFESLRTR TAEALNLDAA LFELIKRCQQ MC SFASQFN SSVSELELRL LQRVDTGLEH PIGSSEWLLS AHKQLTQDMS TQRAIQTEKE QQIETVCETI QNLVDNIKTV LTG HNRQLG DVKHLLKAMA KDEEAALADG EDVPYENSVR QFLGEYKSWQ DNIQTVLFTL VQAMGQVRSQ EHVEMLQEIT PTLK ELKTQ SQSIYNNLVS FASPLVTDAT NECSSPTSSA TYQPSFAAAV RSNTGQKTQP DVMSQNARKL IQKNLATSAD TPPST VPGT GKSVACSPKK AVRDPKTGKA VQERNSYAVS VWKRVKAKLE GRDVDPNRRM SVAEQVDYVI KEATNLDNLA QLYEGW TAW V UniProtKB: Serine/threonine-protein kinase SMG1, Serine/threonine-protein kinase SMG1, Serine/threonine-protein kinase SMG1, Serine/threonine-protein kinase SMG1 |
-Macromolecule #2: INOSITOL HEXAKISPHOSPHATE
| Macromolecule | Name: INOSITOL HEXAKISPHOSPHATE / type: ligand / ID: 2 / Number of copies: 1 / Formula: IHP |
|---|---|
| Molecular weight | Theoretical: 660.035 Da |
| Chemical component information |  ChemComp-IHP: |
-Macromolecule #3: 1-[4-[4-[2-[[4-chloranyl-3-(diethylsulfamoyl)phenyl]amino]pyrimid...
| Macromolecule | Name: 1-[4-[4-[2-[[4-chloranyl-3-(diethylsulfamoyl)phenyl]amino]pyrimidin-4-yl]pyridin-2-yl]phenyl]-3-methyl-urea type: ligand / ID: 3 / Number of copies: 1 / Formula: 88C |
|---|---|
| Molecular weight | Theoretical: 566.074 Da |
| Chemical component information | 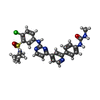 ChemComp-88C: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 89.32 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















