[English] 日本語
 Yorodumi
Yorodumi- EMDB-16454: Electron cryo-tomography of HeLa cells expressing untagged Cidec -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Electron cryo-tomography of HeLa cells expressing untagged Cidec | ||||||||||||||||||
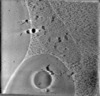
 Map data Map data | Representative tomogram of HeLa cells expressing untagged Cidec | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
| Method | electron tomography / cryo EM | ||||||||||||||||||
 Authors Authors | Ganeva I / Lim K / Boulanger J / Hoffmann PC / Muriel O / Borgeaud AC / Hagen WJH / Savage DB / Kukulski W | ||||||||||||||||||
| Funding support |  United Kingdom, United Kingdom,  Switzerland, 5 items Switzerland, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2023 Journal: Cell Rep / Year: 2023Title: The architecture of Cidec-mediated interfaces between lipid droplets. Authors: Iva Ganeva / Koini Lim / Jerome Boulanger / Patrick C Hoffmann / Olivia Muriel / Alicia C Borgeaud / Wim J H Hagen / David B Savage / Wanda Kukulski /    Abstract: Lipid droplets (LDs) are intracellular organelles responsible for storing surplus energy as neutral lipids. Their size and number vary enormously. In white adipocytes, LDs can reach 100 μm in ...Lipid droplets (LDs) are intracellular organelles responsible for storing surplus energy as neutral lipids. Their size and number vary enormously. In white adipocytes, LDs can reach 100 μm in diameter, occupying >90% of the cell. Cidec, which is strictly required for the formation of large LDs, is concentrated at interfaces between adjacent LDs and facilitates directional flux of neutral lipids from the smaller to the larger LD. The mechanism of lipid transfer is unclear, in part because the architecture of interfaces between LDs remains elusive. Here we visualize interfaces between LDs by electron cryo-tomography and analyze the kinetics of lipid transfer by quantitative live fluorescence microscopy. We show that transfer occurs through closely apposed monolayers, is slowed down by increasing the distance between the monolayers, and follows exponential kinetics. Our data corroborate the notion that Cidec facilitates pressure-driven transfer of neutral lipids through two "leaky" monolayers between LDs. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16454.map.gz emd_16454.map.gz | 14.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16454-v30.xml emd-16454-v30.xml emd-16454.xml emd-16454.xml | 9.7 KB 9.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16454.png emd_16454.png | 205.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16454 http://ftp.pdbj.org/pub/emdb/structures/EMD-16454 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16454 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16454 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16454.map.gz / Format: CCP4 / Size: 21.4 MB / Type: IMAGE STORED AS SIGNED BYTE Download / File: emd_16454.map.gz / Format: CCP4 / Size: 21.4 MB / Type: IMAGE STORED AS SIGNED BYTE | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Representative tomogram of HeLa cells expressing untagged Cidec | ||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 28.3 Å | ||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : HeLa cells expressing untagged Cidec
| Entire | Name: HeLa cells expressing untagged Cidec |
|---|---|
| Components |
|
-Supramolecule #1: HeLa cells expressing untagged Cidec
| Supramolecule | Name: HeLa cells expressing untagged Cidec / type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
| Sectioning | Focused ion beam - Instrument: OTHER / Focused ion beam - Ion: OTHER / Focused ion beam - Voltage: 30 kV / Focused ion beam - Current: 1 nA / Focused ion beam - Duration: 2000 sec. / Focused ion beam - Temperature: 100 K / Focused ion beam - Initial thickness: 1000 nm / Focused ion beam - Final thickness: 200 nm Focused ion beam - Details: Rough milling: 30 kV, 1-0.1 nA. Fine milling: 16kV, 23 pA.. The value given for _em_focused_ion_beam.instrument is FEI Scios. This is not in a list of allowed values ...Focused ion beam - Details: Rough milling: 30 kV, 1-0.1 nA. Fine milling: 16kV, 23 pA.. The value given for _em_focused_ion_beam.instrument is FEI Scios. This is not in a list of allowed values {'OTHER', 'DB235'} so OTHER is written into the XML file. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 5.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Software - Name:  IMOD / Number images used: 121 IMOD / Number images used: 121 |
|---|
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)
















