+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bacteriophage T5 head - pb10-N-Ter fused to OVA | |||||||||
 Map data Map data | To visualize T5 please use 0.0279 contour level To visualize T5 OVA please use 0.0039 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Bacteriophage head Antigen OVA / VIRUS | |||||||||
| Biological species |  Escherichia phage T5 (virus) Escherichia phage T5 (virus) | |||||||||
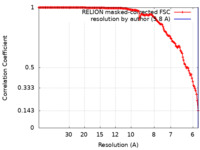
| Method | single particle reconstruction / cryo EM / Resolution: 5.8 Å | |||||||||
 Authors Authors | Schoehn G / Boulanger P / Benihoud K | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: NPJ Vaccines / Year: 2024 Journal: NPJ Vaccines / Year: 2024Title: Antigen self-anchoring onto bacteriophage T5 capsid-like particles for vaccine design. Authors: Emeline Vernhes / Linda Larbi Chérif / Nicolas Ducrot / Clément Vanbergue / Malika Ouldali / Lena Zig / N'diaye Sidibe / Sylviane Hoos / Luis Ramirez-Chamorro / Madalena Renouard / ...Authors: Emeline Vernhes / Linda Larbi Chérif / Nicolas Ducrot / Clément Vanbergue / Malika Ouldali / Lena Zig / N'diaye Sidibe / Sylviane Hoos / Luis Ramirez-Chamorro / Madalena Renouard / Ombeline Rossier / Patrick England / Guy Schoehn / Pascale Boulanger / Karim Benihoud /  Abstract: The promises of vaccines based on virus-like particles stimulate demand for universal non-infectious virus-like platforms that can be efficiently grafted with large antigens. Here, we harnessed the ...The promises of vaccines based on virus-like particles stimulate demand for universal non-infectious virus-like platforms that can be efficiently grafted with large antigens. Here, we harnessed the modularity and extreme affinity of the decoration protein pb10 for the capsid of bacteriophage T5. SPR experiments demonstrated that pb10 fused to mCherry or to the model antigen ovalbumin (Ova) retained picomolar affinity for DNA-free T5 capsid-like particles (T5-CLPs), while cryo-EM studies attested to the full occupancy of the 120 capsid binding sites. Mice immunization with CLP-bound pb10-Ova chimeras elicited strong long-lasting anti-Ova humoral responses involving a large panel of isotypes, as well as CD8 T cell responses, without any extrinsic adjuvant. Therefore, T5-CLP constitutes a unique DNA-free bacteriophage capsid able to display a regular array of large antigens through highly efficient chemical-free anchoring. Its ability to elicit robust immune responses paves the way for further development of this novel vaccination platform. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14863.map.gz emd_14863.map.gz | 225.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14863-v30.xml emd-14863-v30.xml emd-14863.xml emd-14863.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14863_fsc.xml emd_14863_fsc.xml | 14.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_14863.png emd_14863.png | 123 KB | ||
| Filedesc metadata |  emd-14863.cif.gz emd-14863.cif.gz | 4.5 KB | ||
| Others |  emd_14863_half_map_1.map.gz emd_14863_half_map_1.map.gz emd_14863_half_map_2.map.gz emd_14863_half_map_2.map.gz | 226 MB 225.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14863 http://ftp.pdbj.org/pub/emdb/structures/EMD-14863 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14863 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14863 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14863.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14863.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | To visualize T5 please use 0.0279 contour level To visualize T5 OVA please use 0.0039 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.9 Å | ||||||||||||||||||||||||||||||||||||
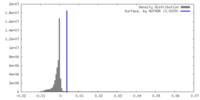
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_14863_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
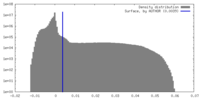
| Density Histograms |
-Half map: #1
| File | emd_14863_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Escherichia phage T5
| Entire | Name:  Escherichia phage T5 (virus) Escherichia phage T5 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia phage T5
| Supramolecule | Name: Escherichia phage T5 / type: virus / ID: 1 / Parent: 0 / Details: E coli; double mutant / NCBI-ID: 2695836 / Sci species name: Escherichia phage T5 / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: Yes |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R3.5/1 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. / Pretreatment - Atmosphere: AIR / Details: 25 mA |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 2-40 / Number grids imaged: 1 / Number real images: 2500 / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.7000000000000001 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)