+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human UPF1 RNA helicase with AMPPNP and RNA | |||||||||
 Map data Map data | Human UPF1 bound to AMPPNP and RNA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA helicase / human / nonsense-mediated mRNA decay / RNA BINDING PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
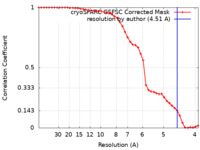
| Method | single particle reconstruction / cryo EM / Resolution: 4.51 Å | |||||||||
 Authors Authors | Langer LM / Conti E | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2024 Journal: Nucleic Acids Res / Year: 2024Title: UPF1 helicase orchestrates mutually exclusive interactions with the SMG6 endonuclease and UPF2. Authors: Lukas M Langer / Katharina Kurscheidt / Jérôme Basquin / Fabien Bonneau / Iuliia Iermak / Claire Basquin / Elena Conti /  Abstract: Nonsense-mediated mRNA decay (NMD) is a conserved co-translational mRNA surveillance and turnover pathway across eukaryotes. NMD has a central role in degrading defective mRNAs and also regulates the ...Nonsense-mediated mRNA decay (NMD) is a conserved co-translational mRNA surveillance and turnover pathway across eukaryotes. NMD has a central role in degrading defective mRNAs and also regulates the stability of a significant portion of the transcriptome. The pathway is organized around UPF1, an RNA helicase that can interact with several NMD-specific factors. In human cells, degradation of the targeted mRNAs begins with a cleavage event that requires the recruitment of the SMG6 endonuclease to UPF1. Previous studies have identified functional links between SMG6 and UPF1, but the underlying molecular mechanisms have remained elusive. Here, we used mass spectrometry, structural biology and biochemical approaches to identify and characterize a conserved short linear motif in SMG6 that interacts with the cysteine/histidine-rich (CH) domain of UPF1. Unexpectedly, we found that the UPF1-SMG6 interaction is precluded when the UPF1 CH domain is engaged with another NMD factor, UPF2. Based on cryo-EM data, we propose that the formation of distinct SMG6-containing and UPF2-containing NMD complexes may be dictated by different conformational states connected to the RNA-binding status of UPF1. Our findings rationalize a key event in metazoan NMD and advance our understanding of mechanisms regulating activity and guiding substrate recognition by the SMG6 endonuclease. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19451.map.gz emd_19451.map.gz | 7.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19451-v30.xml emd-19451-v30.xml emd-19451.xml emd-19451.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19451_fsc.xml emd_19451_fsc.xml | 4.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_19451.png emd_19451.png | 100.7 KB | ||
| Filedesc metadata |  emd-19451.cif.gz emd-19451.cif.gz | 5.2 KB | ||
| Others |  emd_19451_half_map_1.map.gz emd_19451_half_map_1.map.gz emd_19451_half_map_2.map.gz emd_19451_half_map_2.map.gz | 7.4 MB 7.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19451 http://ftp.pdbj.org/pub/emdb/structures/EMD-19451 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19451 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19451 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19451.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19451.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human UPF1 bound to AMPPNP and RNA | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.885 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_19451_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_19451_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : UPF1 bound to AMPPNP
| Entire | Name: UPF1 bound to AMPPNP |
|---|---|
| Components |
|
-Supramolecule #1: UPF1 bound to AMPPNP
| Supramolecule | Name: UPF1 bound to AMPPNP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 124 KDa |
-Macromolecule #1: UPF1, RENT1
| Macromolecule | Name: UPF1, RENT1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSVEAYGPSS QTLTFLDTEE AELLGADTQG SEFEFTDFTL PSQTQTPPGG PGGPGGGGAG GPGGAGAGAA AGQLDAQVGP EGILQNGAVD DSVAKTSQLL AELNFEEDEE DTYYTKDLPI HACSYCGIHD PACVVYCNTS KKWFCNGRGN TSGSHIVNHL VRAKCKEVTL ...String: MSVEAYGPSS QTLTFLDTEE AELLGADTQG SEFEFTDFTL PSQTQTPPGG PGGPGGGGAG GPGGAGAGAA AGQLDAQVGP EGILQNGAVD DSVAKTSQLL AELNFEEDEE DTYYTKDLPI HACSYCGIHD PACVVYCNTS KKWFCNGRGN TSGSHIVNHL VRAKCKEVTL HKDGPLGETV LECYNCGCRN VFLLGFIPAK ADSVVVLLCR QPCASQSSLK DINWDSSQWQ PLIQDRCFLS WLVKIPSEQE QLRARQITAQ QINKLEELWK ENPSATLEDL EKPGVDEEPQ HVLLRYEDAY QYQNIFGPLV KLEADYDKKL KESQTQDNIT VRWDLGLNKK RIAYFTLPKT DSDMRLMQGD EICLRYKGDL APLWKGIGHV IKVPDNYGDE IAIELRSSVG APVEVTHNFQ VDFVWKSTSF DRMQSALKTF AVDETSVSGY IYHKLLGHEV EDVIIKCQLP KRFTAQGLPD LNHSQVYAVK TVLQRPLSLI QGPPGTGKTV TSATIVYHLA RQGNGPVLVC APSNIAVDQL TEKIHQTGLK VVRLCAKSRE AIDSPVSFLA LHNQIRNMDS MPELQKLQQL KDETGELSSA DEKRYRALKR TAERELLMNA DVICCTCVGA GDPRLAKMQF RSILIDESTQ ATEPECMVPV VLGAKQLILV GDHCQLGPVV MCKKAAKAGL SQSLFERLVV LGIRPIRLQV QYRMHPALSA FPSNIFYEGS LQNGVTAADR VKKGFDFQWP QPDKPMFFYV TQGQEEIASS GTSYLNRTEA ANVEKITTKL LKAGAKPDQI GIITPYEGQR SYLVQYMQFS GSLHTKLYQE VEIASVDAFQ GREKDFIILS CVRANEHQGI GFLNDPRRLN VALTRARYGV IIVGNPKALS KQPLWNHLLN YYKEQKVLVE GPLNNLRESL MQFSKPRKLV NTINPGARFM TTAMYDAREA IIPGSVYDRS SQGRPSSMYF QTHDQIGMIS AGPSHVAAMN IPIPFNLVMP PMPPPGYFGQ ANGPAAGRGT PKGKTGRGGR QKNRFGLPGP SQTNLPNSQA SQDVASQPFS QGALTQGYIS MSQPSQMSQP GLSQPELSQD SYLGDEFKSQ IDVALSQDST YQGERAYQHG GVTGLSQY |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 22000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)