-Search query
-Search result
Showing 1 - 50 of 7,573 items for (author: ke & al)

EMDB entry, No image
EMDB-43506: 
Cryo-EM structure of LKB1-STRADalpha-MO25alpha heterocomplex

PDB-8vsu: 
Cryo-EM structure of LKB1-STRADalpha-MO25alpha heterocomplex

EMDB entry, No image
EMDB-40006: 
Hir complex core

EMDB entry, No image
EMDB-40029: 
Hir3 Arm/Tail, Hir2 WD40, Hpc2 C-term

EMDB entry, No image
EMDB-40030: 
Hir1 WD40 domains and Asf1/H3/H4

EMDB entry, No image
EMDB-40037: 
Composite map of the Hir complex with Asf1/H3/H4

EMDB entry, No image
EMDB-40078: 
Chaetomium thermophilum Hir3

EMDB entry, No image
EMDB-43296: 
Constituent EM map: Focused refinement S100A1 of mouse RyR1 in complex with S100A1 (EGTA-only dataset)

EMDB entry, No image
EMDB-43131: 
Voltage gated potassium ion channel Kv1.2 in complex with DTx

EMDB entry, No image
EMDB-43133: 
Voltage gated potassium ion channel Kv1.2 in Sodium

EMDB entry, No image
EMDB-43134: 
Voltage gated potassium ion channel Kv1.2 in Potassium

EMDB entry, No image
EMDB-43136: 
Voltage gated potassium ion channel Kv1.2 W366F, C-type inactivated

PDB-8vc3: 
Voltage gated potassium ion channel Kv1.2 in complex with DTx

PDB-8vc4: 
Voltage gated potassium ion channel Kv1.2 in Sodium

PDB-8vc6: 
Voltage gated potassium ion channel Kv1.2 in Potassium

PDB-8vch: 
Voltage gated potassium ion channel Kv1.2 W366F, C-type inactivated

EMDB entry, No image
EMDB-50580: 
SOLIST cryo-tomogram of native left ventricle mouse heart muscle #1

EMDB entry, No image
EMDB-50582: 
SOLIST native mouse heart muscle tomogram #2

EMDB entry, No image
EMDB-43097: 
Simulation-driven design of prefusion stabilized SARS-CoV-2 spike S2 antigen

EMDB-43527: 
Apoferritin at 100 keV on Alpine detector with a side-entry cryoholder

EMDB-43528: 
Aldolase at 100 keV on the Alpine detector with a side-entry cryoholder

EMDB-17295: 
Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '3 up' RBD conformation

PDB-8oyt: 
Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '3 up' RBD conformation

EMDB-41433: 
Escherichia coli RNA polymerase unwinding intermediate (I1a) at the lambda PR promoter

EMDB-41437: 
Escherichia coli RNA polymerase unwinding intermediate (I1d) at the lambda PR promoter

EMDB-41439: 
Escherichia coli RNA polymerase unwinding intermediate (I1b) at the lambda PR promoter

EMDB-41448: 
Escherichia coli RNA polymerase unwinding intermediate (I1c) at the lambda PR promoter

EMDB-41456: 
Escherichia coli RNA polymerase closed complex intermediate at the lambda PR promoter
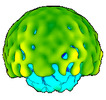
EMDB-50358: 
In vitro-induced genome-releasing intermediate of Rhodobacter microvirus Ebor computed with C5 symmetry

EMDB-19568: 
DtpB hexamer from Streptomyces lividans

PDB-8rwy: 
DtpB hexamer from Streptomyces lividans

EMDB-43088: 
Cryogenic electron microscopy structure of human serum albumin in complex with teniposide

EMDB-43089: 
Cryogenic electron microscopy structure of human serum albumin in complex with salicylic acid

EMDB-43090: 
Cryogenic electron microscopy structure of apo human serum albumin

PDB-8vac: 
Cryogenic electron microscopy structure of human serum albumin in complex with teniposide

PDB-8vae: 
Cryogenic electron microscopy structure of human serum albumin in complex with salicylic acid

PDB-8vaf: 
Cryogenic electron microscopy structure of apo human serum albumin

EMDB-19929: 
Structural basis of D9-THC analog activity at the Cannabinoid 1 receptor

PDB-9erx: 
Structural basis of D9-THC analog activity at the Cannabinoid 1 receptor

EMDB-50296: 
70S Escherichia coli ribosome with P-site initiatior tRNA.

PDB-9fbv: 
70S Escherichia coli ribosome with P-site initiatior tRNA.

EMDB-18592: 
E.coli DNA gyrase in complex with 217 bp substrate DNA and LEI-800

PDB-8qqi: 
E.coli DNA gyrase in complex with 217 bp substrate DNA and LEI-800
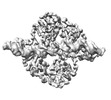
EMDB-18864: 
Central glycolytic genes regulator (CggR) bound to DNA operator

PDB-8r3g: 
Central glycolytic genes regulator (CggR) bound to DNA operator

EMDB-19163: 
Trimeric HSV-1F gB ectodomain in postfusion conformation with three bound HDIT101 Fab molecules.

EMDB-19164: 
Trimeric HSV-1F gB ectodomain in postfusion conformation with three bound HDIT102 Fab molecules.

EMDB-19165: 
Trimeric HSV-2F gB ectodomain in postfusion conformation with three bound HDIT101 Fab molecules.

EMDB-19166: 
Trimeric HSV-2G gB ectodomain in postfusion conformation with three bound HDIT102 Fab molecules.

PDB-8rgz: 
Trimeric HSV-1F gB ectodomain in postfusion conformation with three bound HDIT101 Fab molecules.
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
