[English] 日本語
 Yorodumi
Yorodumi- EMDB-17295: Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '3 up' RBD... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '3 up' RBD conformation | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nanobody / Complex / SARS-CoV-2 / spike / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationvirion component / symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...virion component / symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |    Tequatrovirus T4 Tequatrovirus T4 | |||||||||
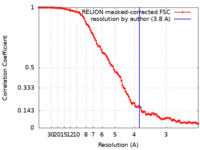
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Weckener M / Naismith JH / Owens RJ | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Open Biol / Year: 2024 Journal: Open Biol / Year: 2024Title: Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2. Authors: Katy Cornish / Jiandong Huo / Luke Jones / Parul Sharma / Joseph W Thrush / Sahar Abdelkarim / Anja Kipar / Siva Ramadurai / Miriam Weckener / Halina Mikolajek / Sai Liu / Imogen Buckle / ...Authors: Katy Cornish / Jiandong Huo / Luke Jones / Parul Sharma / Joseph W Thrush / Sahar Abdelkarim / Anja Kipar / Siva Ramadurai / Miriam Weckener / Halina Mikolajek / Sai Liu / Imogen Buckle / Eleanor Bentley / Adam Kirby / Ximeng Han / Stephen M Laidlaw / Michelle Hill / Lauren Eyssen / Chelsea Norman / Audrey Le Bas / John Clarke / William James / James P Stewart / Miles Carroll / James H Naismith / Raymond J Owens /   Abstract: The Omicron strains of SARS-CoV-2 pose a significant challenge to the development of effective antibody-based treatments as immune evasion has compromised most available immune therapeutics. ...The Omicron strains of SARS-CoV-2 pose a significant challenge to the development of effective antibody-based treatments as immune evasion has compromised most available immune therapeutics. Therefore, in the 'arms race' with the virus, there is a continuing need to identify new biologics for the prevention or treatment of SARS-CoV-2 infections. Here, we report the isolation of nanobodies that bind to the Omicron BA.1 spike protein by screening nanobody phage display libraries previously generated from llamas immunized with either the Wuhan or Beta spike proteins. The structure and binding properties of three of these nanobodies (A8, H6 and B5-5) have been characterized in detail providing insight into their binding epitopes on the Omicron spike protein. Trimeric versions of H6 and B5-5 neutralized the SARS-CoV-2 variant of concern BA.5 both and in the hamster model of COVID-19 following nasal administration. Thus, either alone or in combination could serve as starting points for the development of new anti-viral immunotherapeutics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17295.map.gz emd_17295.map.gz | 9.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17295-v30.xml emd-17295-v30.xml emd-17295.xml emd-17295.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17295_fsc.xml emd_17295_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_17295.png emd_17295.png | 211.7 KB | ||
| Filedesc metadata |  emd-17295.cif.gz emd-17295.cif.gz | 7.2 KB | ||
| Others |  emd_17295_additional_1.map.gz emd_17295_additional_1.map.gz emd_17295_half_map_1.map.gz emd_17295_half_map_1.map.gz emd_17295_half_map_2.map.gz emd_17295_half_map_2.map.gz | 80.8 MB 81 MB 80.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17295 http://ftp.pdbj.org/pub/emdb/structures/EMD-17295 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17295 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17295 | HTTPS FTP |
-Related structure data
| Related structure data |  8oytMC  8owtC  8owvC  8owwC  8oyuC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17295.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17295.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: refined unsharpened map
| File | emd_17295_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | refined unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
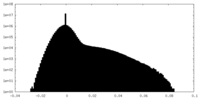
| Density Histograms |
-Half map: half1 map
| File | emd_17295_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half1 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
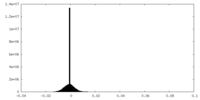
| Density Histograms |
-Half map: half2 map
| File | emd_17295_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half2 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
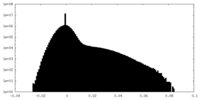
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of stabilised HexaPro Omicron BA.1 spike trimer with thre...
| Entire | Name: Complex of stabilised HexaPro Omicron BA.1 spike trimer with three H6 nanobodies |
|---|---|
| Components |
|
-Supramolecule #1: Complex of stabilised HexaPro Omicron BA.1 spike trimer with thre...
| Supramolecule | Name: Complex of stabilised HexaPro Omicron BA.1 spike trimer with three H6 nanobodies type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|
-Supramolecule #2: H6 nanobody
| Supramolecule | Name: H6 nanobody / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Stabilised HexaPro Omicron BA.1 spike trimer
| Supramolecule | Name: Stabilised HexaPro Omicron BA.1 spike trimer / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Spike glycoprotein,Fibritin
| Macromolecule | Name: Spike glycoprotein,Fibritin / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Tequatrovirus T4 Tequatrovirus T4 |
| Molecular weight | Theoretical: 140.083672 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHVIHV SGTNGTKRFD NPVLPFNDG VYFASIEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLDVYHKNNK SWMESEFRVY S SANNCTFE ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHVIHV SGTNGTKRFD NPVLPFNDG VYFASIEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLDVYHKNNK SWMESEFRVY S SANNCTFE YVSQPFLMDL EGKQGNFKNL REFVFKNIDG YFKIYSKHTP INIVREPEDL PQGFSALEPL VDLPIGINIT RF QTLLALH RSYLTPGDSS SGWTAGAAAY YVGYLQPRTF LLKYNENGTI TDAVDCALDP LSETKCTLKS FTVEKGIYQT SNF RVQPTE SIVRFPNITN LCPFDEVFNA TRFASVYAWN RKRISNCVAD YSVLYNLAPF FTFKCYGVSP TKLNDLCFTN VYAD SFVIR GDEVRQIAPG QTGNIADYNY KLPDDFTGCV IAWNSNKLDS KVSGNYNYLY RLFRKSNLKP FERDISTEIY QAGNK PCNG VAGFNCYFPL KSYSFRPTYG VGHQPYRVVV LSFELLHAPA TVCGPKKSTN LVKNKCVNFN FNGLKGTGVL TESNKK FLP FQQFGRDIAD TTDAVRDPQT LEILDITPCS FGGVSVITPG TNTSNQVAVL YQGVNCTEVP VAIHADQLTP TWRVYST GS NVFQTRAGCL IGAEYVNNSY ECDIPIGAGI CASYQTQTKS HGSASSVASQ SIIAYTMSLG AENSVAYSNN SIAIPTNF T ISVTTEILPV SMTKTSVDCT MYICGDSTEC SNLLLQYGSF CTQLKRALTG IAVEQDKNTQ EVFAQVKQIY KTPPIKYFG GFNFSQILPD PSKPSKRSPI EDLLFNKVTL ADAGFIKQYG DCLGDIAARD LICAQKFKGL TVLPPLLTDE MIAQYTSALL AGTITSGWT FGAGPALQIP FPMQMAYRFN GIGVTQNVLY ENQKLIANQF NSAIGKIQDS LSSTPSALGK LQDVVNHNAQ A LNTLVKQL SSKFGAISSV LNDIFSRLDP PEAEVQIDRL ITGRLQSLQT YVTQQLIRAA EIRASANLAA TKMSECVLGQ SK RVDFCGK GYHLMSFPQS APHGVVFLHV TYVPAQEKNF TTAPAICHDG KAHFPREGVF VSNGTHWFVT QRNFYEPQII TTD NTFVSG NCDVVIGIVN NTVYDPLQPE LDSFKEELDK YFKNHTSPDV DLGDISGINA SVVNIQKEID RLNEVAKNLN ESLI DLQEL GKYEQGSGYI PEAPRDGQAY VRKDGEWVLL STFLGRSLEV LFQGPGHHHH HHHH UniProtKB: Spike glycoprotein, Fibritin |
-Macromolecule #2: H6 nanobody
| Macromolecule | Name: H6 nanobody / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.588144 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLVESGGG LVQPGGSLTL SCVASESSLA PYRVAWFRQA PGKEREGVSC ISRDAHPTST YYTASVKGRF TMSRDNAKNT VYLQMNSLK PSDTAVYYCA TDLGGYCSDS NYPRAWWGQG TQVTVSSKHH HHHH |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 9 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: PBS |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.5 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)