[English] 日本語
 Yorodumi
Yorodumi- EMDB-19164: Trimeric HSV-1F gB ectodomain in postfusion conformation with thr... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Trimeric HSV-1F gB ectodomain in postfusion conformation with three bound HDIT102 Fab molecules. | |||||||||
 Map data Map data | final sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ectodomain / post-fusion / fab molecule / trimeric / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell Golgi membrane / host cell endosome membrane / receptor ligand activity / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Human alphaherpesvirus 1 (Herpes simplex virus type 1) Human alphaherpesvirus 1 (Herpes simplex virus type 1) | |||||||||
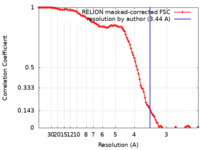
| Method | single particle reconstruction / cryo EM / Resolution: 3.44 Å | |||||||||
 Authors Authors | Kalbermatter D / Seyfizadeh N / Imhof T / Ries M / Mueller C / Jenner L / Blumenschein E / Yendrzheyevskiy A / Moog K / Eckert D ...Kalbermatter D / Seyfizadeh N / Imhof T / Ries M / Mueller C / Jenner L / Blumenschein E / Yendrzheyevskiy A / Moog K / Eckert D / Engel R / Diebolder P / Chami M / Krauss J / Schaller T / Arndt M | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: J Biomed Sci / Year: 2024 Journal: J Biomed Sci / Year: 2024Title: Development of a highly effective combination monoclonal antibody therapy against Herpes simplex virus. Authors: Narges Seyfizadeh / David Kalbermatter / Thomas Imhof / Moritz Ries / Christian Müller / Leonie Jenner / Elisabeth Blumenschein / Alexandra Yendrzheyevskiy / Frank Grün / Kevin Moog / ...Authors: Narges Seyfizadeh / David Kalbermatter / Thomas Imhof / Moritz Ries / Christian Müller / Leonie Jenner / Elisabeth Blumenschein / Alexandra Yendrzheyevskiy / Frank Grün / Kevin Moog / Daniel Eckert / Ronja Engel / Philipp Diebolder / Mohamed Chami / Jürgen Krauss / Torsten Schaller / Michaela Arndt /   Abstract: BACKGROUND: Infections with Herpes simplex virus (HSV)-1 or -2 usually present as mild chronic recurrent disease, however in rare cases can result in life-threatening conditions with a large spectrum ...BACKGROUND: Infections with Herpes simplex virus (HSV)-1 or -2 usually present as mild chronic recurrent disease, however in rare cases can result in life-threatening conditions with a large spectrum of pathology. Monoclonal antibody therapy has great potential especially to treat infections with virus resistant to standard therapies. HDIT101, a humanized IgG targeting HSV-1/2 gB was previously investigated in phase 2 clinical trials. The aim of this study was to develop a next-generation therapy by combining different antiviral monoclonal antibodies. METHODS: A lymph-node derived phage display library (LYNDAL) was screened against recombinant gB from Herpes simplex virus (HSV) -1 and HDIT102 scFv was selected for its binding characteristics using ...METHODS: A lymph-node derived phage display library (LYNDAL) was screened against recombinant gB from Herpes simplex virus (HSV) -1 and HDIT102 scFv was selected for its binding characteristics using bio-layer interferometry. HDIT102 was further developed as fully human IgG and tested alone or in combination with HDIT101, a clinically tested humanized anti-HSV IgG, in vitro and in vivo. T-cell stimulating activities by antigen-presenting cells treated with IgG-HSV immune complexes were analyzed using primary human cells. To determine the epitopes, the cryo-EM structures of HDIT101 or HDIT102 Fab bound to HSV-1F as well as HSV-2G gB protein were solved at resolutions < 3.5 Å. RESULTS: HDIT102 Fab showed strong binding to HSV-1F gB with Kd of 8.95 × 10 M and to HSV-2G gB with Kd of 3.29 × 10 M. Neutralization of cell-free virus and inhibition of cell-to-cell ...RESULTS: HDIT102 Fab showed strong binding to HSV-1F gB with Kd of 8.95 × 10 M and to HSV-2G gB with Kd of 3.29 × 10 M. Neutralization of cell-free virus and inhibition of cell-to-cell spread were comparable between HDIT101 and HDIT102. Both antibodies induced internalization of gB from the cell surface into acidic endosomes by binding distinct epitopes in domain I of gB and compete for binding. CryoEM analyses revealed the ability to form heterogenic immune complexes consisting of two HDIT102 and one HDIT101 Fab bound to one gB trimeric molecule. Both antibodies mediated antibody-dependent phagocytosis by antigen presenting cells which stimulated autologous T-cell activation. In vivo, the combination of HDIT101 and HDIT102 demonstrated synergistic effects on survival and clinical outcome in immunocompetent BALB/cOlaHsd mice. CONCLUSION: This biochemical and immunological study showcases the potential of an effective combination therapy with two monoclonal anti-gB IgGs for the treatment of HSV-1/2 induced disease conditions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19164.map.gz emd_19164.map.gz | 9.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19164-v30.xml emd-19164-v30.xml emd-19164.xml emd-19164.xml | 22 KB 22 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19164_fsc.xml emd_19164_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_19164.png emd_19164.png | 47.6 KB | ||
| Masks |  emd_19164_msk_1.map emd_19164_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19164.cif.gz emd-19164.cif.gz | 7.2 KB | ||
| Others |  emd_19164_half_map_1.map.gz emd_19164_half_map_1.map.gz emd_19164_half_map_2.map.gz emd_19164_half_map_2.map.gz | 81 MB 80.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19164 http://ftp.pdbj.org/pub/emdb/structures/EMD-19164 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19164 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19164 | HTTPS FTP |
-Related structure data
| Related structure data |  8rh0MC  8rgzC  8rh1C  8rh2C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19164.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19164.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | final sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2028 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19164_msk_1.map emd_19164_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_19164_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_19164_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Trimeric HSV-1F gB ectodomain in post-fusion conformation with th...
| Entire | Name: Trimeric HSV-1F gB ectodomain in post-fusion conformation with three bound HDIT102 Fab molecules |
|---|---|
| Components |
|
-Supramolecule #1: Trimeric HSV-1F gB ectodomain in post-fusion conformation with th...
| Supramolecule | Name: Trimeric HSV-1F gB ectodomain in post-fusion conformation with three bound HDIT102 Fab molecules type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 523 KDa |
-Macromolecule #1: Envelope glycoprotein B
| Macromolecule | Name: Envelope glycoprotein B / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human alphaherpesvirus 1 (Herpes simplex virus type 1) Human alphaherpesvirus 1 (Herpes simplex virus type 1) |
| Molecular weight | Theoretical: 100.449375 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRQGAPARGC RWFVVWALLG LTLGVLVASA APSSPGTPGV AAATQAANGG PATPAPPAPG PAPTGDTKPK KNKKPKNPPP PRPAGDNAT VAAGHATLRE HLRDIKAENT DANFYVCPPP TGATVVQFEQ PRRCPTRPEG QNYTEGIAVV FKENIAPYKF K ATMYYKDV ...String: MRQGAPARGC RWFVVWALLG LTLGVLVASA APSSPGTPGV AAATQAANGG PATPAPPAPG PAPTGDTKPK KNKKPKNPPP PRPAGDNAT VAAGHATLRE HLRDIKAENT DANFYVCPPP TGATVVQFEQ PRRCPTRPEG QNYTEGIAVV FKENIAPYKF K ATMYYKDV TVSQVWFGHR YSQFMGIFED RAPVPFEEVI DKINAKGVCR STAKYVRNNL ETTAFHRDDH ETDMELKPAN AA TRTSRGW HTTDLKYNPS RVEAFHRYGT TVNCIVEEVD ARSVYPYDEF VLATGDFVYM SPFYGYREGS HTEHTSYAAD RFK QVDGFY ARDLTTKARA TAPTTRNLLT TPKFTVAWDW VPKRPSVCTM TKWQEVDEML RSEYGGSFRF SSDAISTTFT TNLT EYPLS RVDLGDCIGK DARDAMDRIF ARRYNATHIK VGQPQYYLAN GGFLIAYQPL LSNTLAELYV REHLREQSRK PPNPT PPPP GASANASVER IKTTSSIEFA RLQFTYNHIQ RHVNDMLGRV AIAWCELQNH ELTLWNEARK LNPNAIASAT VGRRVS ARM LGDVMAVSTC VPVAADNVIV QNSMRISSRP GACYSRPLVS FRYEDQGPLV EGQLGENNEL RLTRDAIEPC TVGHRRY FT FGGGYVYFEE YAYSHQLSRA DITTVSTFID LNITMLEDHE FVPLEVYTRH EIKDSGLLDY TEVQRRNQLH DLRFADID T VIHADANAAM FAGLGAFFEG MGDLGRAVGK VVMGIVGGVV SAVSGVSSFM SNPFGALAVG LLVLAGLAAA FFAFRYVMR LQSNPMKALY PLTTKELKNP TNPDASGEGE EGGDFDEAKL AEAREMIRYM ALVSAMEHTE HKAKKKGTSA LLSAKVTDMV MRKRRNTNY TQVPNKDGDA DEDDLAA UniProtKB: Envelope glycoprotein B |
-Macromolecule #2: HDIT102 Fab heavy chain
| Macromolecule | Name: HDIT102 Fab heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 49.310387 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVQSGAE VKTPGASVRV SCKASGHTFR TFDINWVRQA AGQGLEWMGW MSPNSGNTGY ARQFQGRVTM TRNISANTAY MELRGLRFD DTAVYYCARG PGSTGTTGSM DVWGQGTTVT VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV T VSWNSGAL ...String: EVQLVQSGAE VKTPGASVRV SCKASGHTFR TFDINWVRQA AGQGLEWMGW MSPNSGNTGY ARQFQGRVTM TRNISANTAY MELRGLRFD DTAVYYCARG PGSTGTTGSM DVWGQGTTVT VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV T VSWNSGAL TSGVHTFPAV LQSSGLYSLS SVVTVPSSSL GTQTYICNVN HKPSNTKVDK KVEPKSCDKT HTCPPCPAPE LL GGPSVFL FPPKPKDTLM ISRTPEVTCV VVDVSHEDPE VKFNWYVDGV EVHNAKTKPR EEQYNSTYRV VSVLTVLHQD WLN GKEYKC KVSNKALPAP IEKTISKAKG QPREPQVYTL PPSRDELTKN QVSLTCLVKG FYPSDIAVEW ESNGQPENNY KTTP PVLDS DGSFFLYSKL TVDKSRWQQG NVFSCSVMHE ALHNHYTQKS LSLSPGK |
-Macromolecule #3: HDIT102 Fab heavy chain
| Macromolecule | Name: HDIT102 Fab heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.403775 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QAGLTQPPSV SVAPGKTARI SCGGNNIGSK SVHWYQQKPG QAPVLVIYYD SDRPSGIPER FSGSNSGNTA TLTISRVEAG DEADYYCQV WDSGSVVFGG GTKLTVLGQP KAAPSVTLFP PSSEELQANK ATLVCLISDF YPGAVTVAWK ADSSPVKAGV E TTTPSKQS ...String: QAGLTQPPSV SVAPGKTARI SCGGNNIGSK SVHWYQQKPG QAPVLVIYYD SDRPSGIPER FSGSNSGNTA TLTISRVEAG DEADYYCQV WDSGSVVFGG GTKLTVLGQP KAAPSVTLFP PSSEELQANK ATLVCLISDF YPGAVTVAWK ADSSPVKAGV E TTTPSKQS NNKYAASSYL SLTPEQWKSH RSYSCQVTHE GSTVEKTVAP TECS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 283.15 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 6611 / Average exposure time: 4.0 sec. / Average electron dose: 1.15 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)