+Search query
-Structure paper
| Title | Structure of the Hir histone chaperone complex. |
|---|---|
| Journal, issue, pages | Mol Cell, Vol. 84, Issue 14, Page 2601-2617.e12, Year 2024 |
| Publish date | Jul 25, 2024 |
 Authors Authors | Hee Jong Kim / Mary R Szurgot / Trevor van Eeuwen / M Daniel Ricketts / Pratik Basnet / Athena L Zhang / Austin Vogt / Samah Sharmin / Craig D Kaplan / Benjamin A Garcia / Ronen Marmorstein / Kenji Murakami /  |
| PubMed Abstract | The evolutionarily conserved HIRA/Hir histone chaperone complex and ASF1a/Asf1 co-chaperone cooperate to deposit histone (H3/H4) tetramers on DNA for replication-independent chromatin assembly. The ...The evolutionarily conserved HIRA/Hir histone chaperone complex and ASF1a/Asf1 co-chaperone cooperate to deposit histone (H3/H4) tetramers on DNA for replication-independent chromatin assembly. The molecular architecture of the HIRA/Hir complex and its mode of histone deposition have remained unknown. Here, we report the cryo-EM structure of the S. cerevisiae Hir complex with Asf1/H3/H4 at 2.9-6.8 Å resolution. We find that the Hir complex forms an arc-shaped dimer with a Hir1/Hir2/Hir3/Hpc2 stoichiometry of 2/4/2/4. The core of the complex containing two Hir1/Hir2/Hir2 trimers and N-terminal segments of Hir3 forms a central cavity containing two copies of Hpc2, with one engaged by Asf1/H3/H4, in a suitable position to accommodate a histone (H3/H4) tetramer, while the C-terminal segments of Hir3 harbor nucleic acid binding activity to wrap DNA around the Hpc2-assisted histone tetramer. The structure suggests a model for how the Hir/Asf1 complex promotes the formation of histone tetramers for their subsequent deposition onto DNA. |
 External links External links |  Mol Cell / Mol Cell /  PubMed:38925115 / PubMed:38925115 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.96 - 12 Å |
| Structure data | EMDB-40006: Hir complex core EMDB-40029: Hir3 Arm/Tail, Hir2 WD40, Hpc2 C-term EMDB-40030, PDB-8ghm: EMDB-40037: Composite map of the Hir complex with Asf1/H3/H4 EMDB-40078, PDB-8gix: |
| Chemicals | 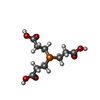 ChemComp-TCE: |
| Source |
|
 Keywords Keywords | CHAPERONE / Histone / Complex / Replication-Independent / subunit |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers














 Thermochaetoides thermophila (fungus)
Thermochaetoides thermophila (fungus)