-Search query
-Search result
Showing 1 - 50 of 236 items for (author: jakob & r)

EMDB-18973: 
Cryo-EM structure of Human SHMT1

PDB-8r7h: 
Cryo-EM structure of Human SHMT1

EMDB-19426: 
Escherichia coli 50S subunit in complex with the antimicrobial peptide Api137

EMDB-19427: 
Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation I

EMDB-19428: 
Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation II

EMDB-19429: 
Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation III

PDB-8rpy: 
Escherichia coli 50S subunit in complex with the antimicrobial peptide Api137

PDB-8rpz: 
Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation I

PDB-8rq0: 
Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation II

PDB-8rq2: 
Escherichia coli 50S subunit in complex with the antimicrobial peptide Api88 - conformation III
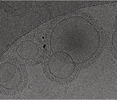
EMDB-16813: 
Tomogram of GBP1 coatomers assembled on brain polar lipid-derived small unilamellar vesicles.

EMDB-16814: 
Tomogram of GBP1 coatomers assembled on brain polar lipid-derived small unilamellar vesicles.

EMDB-16815: 
Tomogram of GBP1 coatomers assembled on brain polar lipid-derived small unilamellar vesicles.

EMDB-16794: 
Cryo-EM structure of the human GBP1 dimer bound to GDP-AlF3

EMDB-18728: 
Rat synaptosome
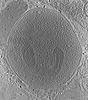
EMDB-18744: 
Stimulated rat synaptosome.

EMDB-18746: 
wild type neuronal synapse

EMDB-18748: 
SNAP-25-4E neuronal synapse

EMDB-18749: 
SNAP-25-4K neuronal synapse

EMDB-41667: 
Cryo-EM structure of the PP2A:B55-FAM122A complex, B55 body

EMDB-41668: 
Cryo-EM structure of the PP2A:B55-FAM122A complex, PP2Ac body

PDB-8twe: 
Cryo-EM structure of the PP2A:B55-FAM122A complex, B55 body

PDB-8twi: 
Cryo-EM structure of the PP2A:B55-FAM122A complex, PP2Ac body

EMDB-40644: 
Cryo-EM structure of the PP2A:B55-FAM122A complex

EMDB-41604: 
Cryo-EM structure of the PP2A:B55-ARPP19 complex

PDB-8so0: 
Cryo-EM structure of the PP2A:B55-FAM122A complex

PDB-8ttb: 
Cryo-EM structure of the PP2A:B55-ARPP19 complex

EMDB-15480: 
Subtomogram average of nucleosomes extracted from euchromatin and facultative heterochromatin nanodomains of Drosophila melanogaster embryos

EMDB-15481: 
Subtomogram average of nucleosomes extracted from euchromatin and facultative heterochromatin nanodomains of Drosophila melanogaster embryos

EMDB-15483: 
Subtomogram average of nucleosomes extracted from constitutive heterochromatin of Drosophila melanogaster embryos
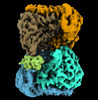
EMDB-15065: 
Cryo-EM structure of the Human SHMT1-RNA complex

PDB-8a11: 
Cryo-EM structure of the Human SHMT1-RNA complex

EMDB-15274: 
Single Particle cryo-EM of the lipid binding protein P116 (MPN213) from Mycoplasma pneumoniae at 3.3 Angstrom resolution.

EMDB-15275: 
Single Particle cryo-EM of the empty lipid binding protein P116 (MPN213) from Mycoplasma pneumoniae at 4 Angstrom resolution

EMDB-15276: 
Single Particle cryo-EM of the lipid binding protein P116 (MPN213) refilled with FBS from Mycoplasma pneumoniae at 3.5 Angstrom resolution.

EMDB-15277: 
Single Particle cryo-EM of the lipid binding protein P116 (MPN213) from Mycoplasma pneumoniae bound to HDL.

PDB-8a9a: 
Single Particle cryo-EM of the lipid binding protein P116 (MPN213) from Mycoplasma pneumoniae at 3.3 Angstrom resolution.

PDB-8a9b: 
Single Particle cryo-EM of the empty lipid binding protein P116 (MPN213) from Mycoplasma pneumoniae at 4 Angstrom resolution
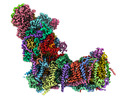
EMDB-14791: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (inhibited by DDM)

EMDB-14792: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (inhibited by DDM) - membrane arm
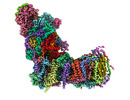
EMDB-14794: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 2)
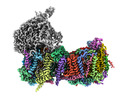
EMDB-14796: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 2) - membrane arm
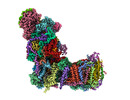
EMDB-14797: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 1)
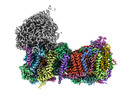
EMDB-14798: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 1) - membrane arm

PDB-7zm7: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (inhibited by DDM)

PDB-7zm8: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (inhibited by DDM) - membrane arm

PDB-7zmb: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 2)

PDB-7zme: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 2) - membrane arm

PDB-7zmg: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 1)

PDB-7zmh: 
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 1) - membrane arm
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
