[English] 日本語
 Yorodumi
Yorodumi- EMDB-16814: Tomogram of GBP1 coatomers assembled on brain polar lipid-derived... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 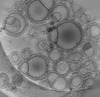 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tomogram of GBP1 coatomers assembled on brain polar lipid-derived small unilamellar vesicles. | |||||||||
 Map data Map data | Electron cryotomogram of membrane-associated GBP1 coatomers on BPLE SUVs. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GBP1 / cryo-ET / liposome / coatomer / IMMUNE SYSTEM | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Kuhm TI / Jakobi AJ | |||||||||
| Funding support | European Union,  Netherlands, 2 items Netherlands, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2025 Journal: Nat Struct Mol Biol / Year: 2025Title: Structural basis of antimicrobial membrane coat assembly by human GBP1. Authors: Tanja Kuhm / Clémence Taisne / Cecilia de Agrela Pinto / Luca Gross / Evdokia A Giannopoulou / Stefan T Huber / Els Pardon / Jan Steyaert / Sander J Tans / Arjen J Jakobi /   Abstract: Guanylate-binding proteins (GBPs) are interferon-inducible guanosine triphosphate hydrolases (GTPases) mediating host defense against intracellular pathogens. Their antimicrobial activity hinges on ...Guanylate-binding proteins (GBPs) are interferon-inducible guanosine triphosphate hydrolases (GTPases) mediating host defense against intracellular pathogens. Their antimicrobial activity hinges on their ability to self-associate and coat pathogen-associated compartments or cytosolic bacteria. Coat formation depends on GTPase activity but how nucleotide binding and hydrolysis prime coat formation remains unclear. Here, we report the cryo-electron microscopy structure of the full-length human GBP1 dimer in its guanine nucleotide-bound state and describe the molecular ultrastructure of the GBP1 coat on liposomes and bacterial lipopolysaccharide membranes. Conformational changes of the middle and GTPase effector domains expose the isoprenylated C terminus for membrane association. The α-helical middle domains form a parallel, crossover arrangement essential for coat formation and position the extended effector domain for intercalation into the lipopolysaccharide layer of gram-negative membranes. Nucleotide binding and hydrolysis create oligomeric scaffolds with contractile abilities that promote membrane extrusion and fragmentation. Our data offer a structural and mechanistic framework for understanding GBP1 effector functions in intracellular immunity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16814.map.gz emd_16814.map.gz | 3.2 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16814-v30.xml emd-16814-v30.xml emd-16814.xml emd-16814.xml | 12.3 KB 12.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16814.png emd_16814.png | 180.1 KB | ||
| Filedesc metadata |  emd-16814.cif.gz emd-16814.cif.gz | 4.3 KB | ||
| Others |  emd_16814_additional_1.map.gz emd_16814_additional_1.map.gz | 25.4 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16814 http://ftp.pdbj.org/pub/emdb/structures/EMD-16814 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16814 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16814 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16814.map.gz / Format: CCP4 / Size: 3.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16814.map.gz / Format: CCP4 / Size: 3.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Electron cryotomogram of membrane-associated GBP1 coatomers on BPLE SUVs. | ||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 6.15 Å | ||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Electron cryotomogram of membrane-associated GBP1 coatomers on BPLE...
| File | emd_16814_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Electron cryotomogram of membrane-associated GBP1 coatomers on BPLE SUVs. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Membrane-assembled coatomer formed by GDP-AlF3-stabilised GBP1 di...
| Entire | Name: Membrane-assembled coatomer formed by GDP-AlF3-stabilised GBP1 dimers on brain polar lipid-derived SUVs. |
|---|---|
| Components |
|
-Supramolecule #1: Membrane-assembled coatomer formed by GDP-AlF3-stabilised GBP1 di...
| Supramolecule | Name: Membrane-assembled coatomer formed by GDP-AlF3-stabilised GBP1 dimers on brain polar lipid-derived SUVs. type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 98 % / Chamber temperature: 20 K / Instrument: LEICA EM GP / Details: Blotted for 4 seconds from the carbon side.. |
| Sectioning | Other: NO SECTIONING |
| Fiducial marker | Manufacturer: CMC Utrecht / Diameter: 10 nm |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FSC |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 61 / Average exposure time: 2.0 sec. / Average electron dose: 1.54 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 12000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 4.1 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 5.0 µm |
| Sample stage | Specimen holder model: JEOL 3200FSC CRYOHOLDER / Cooling holder cryogen: NITROGEN |
- Image processing
Image processing
| Final reconstruction | Algorithm: BACK PROJECTION / Software - Name:  IMOD / Number images used: 62 IMOD / Number images used: 62 |
|---|
 Movie
Movie Controller
Controller





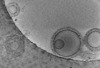

 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)
























