-Search query
-Search result
Showing 1 - 50 of 53 items for (author: huang & cs)
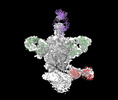
EMDB-44246: 
Cryo-EM structure of HIV-1 JRFL v6 Env in complex with vaccine-elicited, Membrane Proximal External Region (MPER) directed antibody DH1317.4.

EMDB-38216: 
Cryo-EM structure of SARS-CoV-2 S-BQ.1 in complex with antibody O5C2

EMDB-16434: 
Type Ib beta-amyloid 42 Filaments from Human Brain
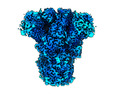
EMDB-26378: 
Cryo-EM structure of PDF-2180 Spike glycoprotein

PDB-7u6r: 
Cryo-EM structure of PDF-2180 Spike glycoprotein

EMDB-27779: 
Structure of the SARS-CoV-2 spike glycoprotein S2 subunit

PDB-8dya: 
Structure of the SARS-CoV-2 spike glycoprotein S2 subunit

EMDB-33248: 
Cryo-EM structure of DHEA-ADGRG2-BT-Gs complex

EMDB-33249: 
Cryo-EM structure of DHEA-ADGRG2-FL-Gs complex

EMDB-33250: 
Cryo-EM structure of DHEA-ADGRG2-BT-Gs complex at lower state

EMDB-14174: 
TMEM106B filaments with Fold I from Alzheimer's disease (case 1)

EMDB-14176: 
TMEM106B filaments with Fold I-d from Multiple system atrophy (case 18)

EMDB-14187: 
TMEM106B filaments with Fold IIa from Multiple system atrophy (case 19)

EMDB-14188: 
TMEM106B filaments with Fold IIb from Multiple system atrophy (case 19)

EMDB-14189: 
TMEM106B filaments with Fold III from Multiple system atrophy (case 17)
PDB-7qvc: 
TMEM106B filaments with Fold I from Alzheimer's disease (case 1)
PDB-7qvf: 
TMEM106B filaments with Fold I-d from Multiple system atrophy (case 18)
PDB-7qwg: 
TMEM106B filaments with Fold IIa from Multiple system atrophy (case 19)
PDB-7qwl: 
TMEM106B filaments with Fold IIb from Multiple system atrophy (case 19)

PDB-7qwm: 
TMEM106B filaments with Fold III from Multiple system atrophy (case 17)

EMDB-13800: 
Type I beta-amyloid 42 Filaments from Human Brain

EMDB-13809: 
Type II beta-amyloid 42 Filaments from Human Brain
PDB-7q4b: 
Type I beta-amyloid 42 Filaments from Human Brain

PDB-7q4m: 
Type II beta-amyloid 42 Filaments from Human Brain

EMDB-22995: 
Spike protein trimer

EMDB-21961: 
Cryo-EM structure of the VRC315 clinical trial, vaccine-elicited, human antibody 1D12 in complex with an H7 SH13 HA trimer

EMDB-22993: 
SARS-CoV-2 spike glycoprotein:Fab 5A6 complex I

EMDB-22994: 
SARS-CoV-2 Spike protein in complex with Fab 2H4

EMDB-22997: 
SARS-CoV-2 spike glycoprotein:Fab 3D11 complex

EMDB-23707: 
SARS-CoV-2 Spike:5A6 Fab complex I focused refinement

EMDB-23709: 
SARS-CoV-2 Spike:Fab 3D11 complex focused refinement

EMDB-30700: 
Human SARM1 inhibitory state bounded with inhibitor dHNN

EMDB-22804: 
Cryo-EM structure of SRR2899884.46167H+MEDI8852L fab in complex with Victoria HA

EMDB-22443: 
Cryo-EM structure of ABCG5/G8 in complex with Fab 2E10 and 11F4

EMDB-21035: 
Structure of NPC1-like intracellular cholesterol transporter 1 (NPC1L1)

EMDB-21037: 
Structure of NPC1-like intracellular cholesterol transporter 1 (NPC1L1) in complex with an ezetimibe analog

EMDB-8977: 
Cryo-EM structure at 3.2 A resolution of HIV-1 fusion peptide-directed antibody, A12V163-b.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122

EMDB-20191: 
Cryo-EM structure of vaccine-elicited antibody 0PV-b.01 in complex with HIV-1 Env BG505 DS-SOSIP and antibodies VRC03 and PGT122

EMDB-20189: 
Cryo-EM structure of vaccine-elicited antibody 0PV-a.01 in complex with HIV-1 Env BG505 DS-SOSIP and antibodies VRC03 and PGT122

EMDB-9189: 
Cryo-EM structure at 3.8 A resolution of HIV-1 fusion peptide-directed antibody, DF1W-a.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122

EMDB-9319: 
Cryo-EM structure at 4.0 A resolution of vaccine-elicited antibody A12V163-a.01 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122

EMDB-9320: 
Cryo-EM structure at 4.2 A resolution of vaccine-elicited antibody DFPH-a.15 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122

EMDB-9359: 
Cryo-EM structure of vaccine-elicited antibody 0PV-c.01 in complex with HIV-1 Env BG505 DS-SOSIP and antibodies VRC03 and PGT122

EMDB-9135: 
Cryo-EM structure of Human Parainfluenza Virus Type 3 (hPIV3) in complex with antibody PIA174

EMDB-7622: 
Cryo-EM structure at 4.2 A resolution of vaccine-elicited antibody vFP1.01 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122

EMDB-7621: 
Cryo-EM structure at 4.0 A resolution of vaccine-elicited antibody vFP7.04 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122

EMDB-7459: 
Cryo-EM structure at 3.8 A resolution of vaccine-elicited antibody vFP20.01 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122

EMDB-7460: 
Cryo-EM structure at 3.6 A resolution of vaccine-elicited antibody vFP16.02 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122

EMDB-8420: 
Cryo-EM structure of BG505 DS-SOSIP HIV-1 Env trimer in complex with vaccine elicited, fusion peptide-directed antibody vFP1.01

EMDB-8421: 
Cryo-EM structure of an asymmetric complex of BG505 DS-SOSIP HIV-1 Env trimer with vaccine elicited, fusion peptide-directed antibody vFP5.01
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
