[English] 日本語
 Yorodumi
Yorodumi- EMDB-14174: TMEM106B filaments with Fold I from Alzheimer's disease (case 1) -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-14174 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | TMEM106B filaments with Fold I from Alzheimer's disease (case 1) | |||||||||
 Map data Map data | Sharpened map of TMEM106B filaments with Fold I from Alzheimer's disease (case 1) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Amyloid / neurodegeneration / filament / TMEM106B / PROTEIN FIBRIL | |||||||||
| Function / homology |  Function and homology information Function and homology informationlysosomal protein catabolic process / lysosomal lumen acidification / regulation of lysosome organization / lysosome localization / positive regulation of dendrite development / lysosomal transport / lysosome organization / dendrite morphogenesis / neuron cellular homeostasis / late endosome membrane ...lysosomal protein catabolic process / lysosomal lumen acidification / regulation of lysosome organization / lysosome localization / positive regulation of dendrite development / lysosomal transport / lysosome organization / dendrite morphogenesis / neuron cellular homeostasis / late endosome membrane / ATPase binding / lysosome / endosome / lysosomal membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
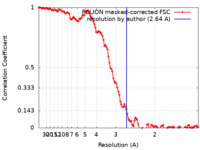
| Method | helical reconstruction / cryo EM / Resolution: 2.64 Å | |||||||||
 Authors Authors | Lovestam S / Schweighauser M | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Age-dependent formation of TMEM106B amyloid filaments in human brains. Authors: Manuel Schweighauser / Diana Arseni / Mehtap Bacioglu / Melissa Huang / Sofia Lövestam / Yang Shi / Yang Yang / Wenjuan Zhang / Abhay Kotecha / Holly J Garringer / Ruben Vidal / Grace I ...Authors: Manuel Schweighauser / Diana Arseni / Mehtap Bacioglu / Melissa Huang / Sofia Lövestam / Yang Shi / Yang Yang / Wenjuan Zhang / Abhay Kotecha / Holly J Garringer / Ruben Vidal / Grace I Hallinan / Kathy L Newell / Airi Tarutani / Shigeo Murayama / Masayuki Miyazaki / Yuko Saito / Mari Yoshida / Kazuko Hasegawa / Tammaryn Lashley / Tamas Revesz / Gabor G Kovacs / John van Swieten / Masaki Takao / Masato Hasegawa / Bernardino Ghetti / Maria Grazia Spillantini / Benjamin Ryskeldi-Falcon / Alexey G Murzin / Michel Goedert / Sjors H W Scheres /       Abstract: Many age-dependent neurodegenerative diseases, such as Alzheimer's and Parkinson's, are characterized by abundant inclusions of amyloid filaments. Filamentous inclusions of the proteins tau, amyloid- ...Many age-dependent neurodegenerative diseases, such as Alzheimer's and Parkinson's, are characterized by abundant inclusions of amyloid filaments. Filamentous inclusions of the proteins tau, amyloid-β, α-synuclein and transactive response DNA-binding protein (TARDBP; also known as TDP-43) are the most common. Here we used structure determination by cryogenic electron microscopy to show that residues 120-254 of the lysosomal type II transmembrane protein 106B (TMEM106B) also form amyloid filaments in human brains. We determined the structures of TMEM106B filaments from a number of brain regions of 22 individuals with abundant amyloid deposits, including those resulting from sporadic and inherited tauopathies, amyloid-β amyloidoses, synucleinopathies and TDP-43 proteinopathies, as well as from the frontal cortex of 3 individuals with normal neurology and no or only a few amyloid deposits. We observed three TMEM106B folds, with no clear relationships between folds and diseases. TMEM106B filaments correlated with the presence of a 29-kDa sarkosyl-insoluble fragment and globular cytoplasmic inclusions, as detected by an antibody specific to the carboxy-terminal region of TMEM106B. The identification of TMEM106B filaments in the brains of older, but not younger, individuals with normal neurology indicates that they form in an age-dependent manner. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14174.map.gz emd_14174.map.gz | 48.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14174-v30.xml emd-14174-v30.xml emd-14174.xml emd-14174.xml | 15.1 KB 15.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14174_fsc.xml emd_14174_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_14174.png emd_14174.png | 37.4 KB | ||
| Filedesc metadata |  emd-14174.cif.gz emd-14174.cif.gz | 5.2 KB | ||
| Others |  emd_14174_half_map_1.map.gz emd_14174_half_map_1.map.gz emd_14174_half_map_2.map.gz emd_14174_half_map_2.map.gz | 98.3 MB 98.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14174 http://ftp.pdbj.org/pub/emdb/structures/EMD-14174 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14174 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14174 | HTTPS FTP |
-Related structure data
| Related structure data |  7qvcMC  7qvfC  7qwgC  7qwlC  7qwmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14174.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14174.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of TMEM106B filaments with Fold I from Alzheimer's disease (case 1) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.727 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
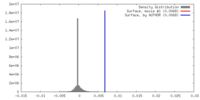
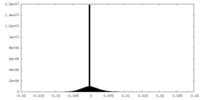
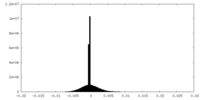
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half map 2 of TMEM106B filaments with Fold...
| File | emd_14174_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 of TMEM106B filaments with Fold I from Alzheimer's disease (case 1) | ||||||||||||
| Projections & Slices |
| ||||||||||||
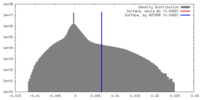
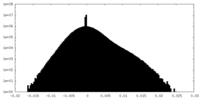
| Density Histograms |
-Half map: Half map 1 of TMEM106B filaments with Fold...
| File | emd_14174_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 of TMEM106B filaments with Fold I from Alzheimer's disease (case 1) | ||||||||||||
| Projections & Slices |
| ||||||||||||
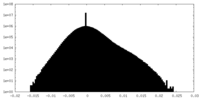
| Density Histograms |
- Sample components
Sample components
-Entire : TMEM106B
| Entire | Name: TMEM106B |
|---|---|
| Components |
|
-Supramolecule #1: TMEM106B
| Supramolecule | Name: TMEM106B / type: tissue / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Transmembrane protein 106B
| Macromolecule | Name: Transmembrane protein 106B / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 31.142295 KDa |
| Sequence | String: MGKSLSHLPL HSSKEDAYDG VTSENMRNGL VNSEVHNEDG RNGDVSQFPY VEFTGRDSVT CPTCQGTGRI PRGQENQLVA LIPYSDQRL RPRRTKLYVM ASVFVCLLLS GLAVFFLFPR SIDVKYIGVK SAYVSYDVQK RTIYLNITNT LNITNNNYYS V EVENITAQ ...String: MGKSLSHLPL HSSKEDAYDG VTSENMRNGL VNSEVHNEDG RNGDVSQFPY VEFTGRDSVT CPTCQGTGRI PRGQENQLVA LIPYSDQRL RPRRTKLYVM ASVFVCLLLS GLAVFFLFPR SIDVKYIGVK SAYVSYDVQK RTIYLNITNT LNITNNNYYS V EVENITAQ VQFSKTVIGK ARLNNISIIG PLDMKQIDYT VPTVIAEEMS YMYDFCTLIS IKVHNIVLMM QVTVTTTYFG HS EQISQER YQYVDCGRNT TYQLGQSEYL NVLQPQQ UniProtKB: Transmembrane protein 106B |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)