+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30700 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human SARM1 inhibitory state bounded with inhibitor dHNN | |||||||||
 Map data Map data | Human SARM1 bounded with inhibitor dHNN | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NAD(+) hydrolase / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of MyD88-independent toll-like receptor signaling pathway / extrinsic component of synaptic membrane / MyD88-independent TLR4 cascade / NADP+ nucleosidase activity / Toll Like Receptor 3 (TLR3) Cascade / NAD+ catabolic process / NAD+ nucleosidase activity / regulation of synapse pruning / modification of postsynaptic structure / ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase ...negative regulation of MyD88-independent toll-like receptor signaling pathway / extrinsic component of synaptic membrane / MyD88-independent TLR4 cascade / NADP+ nucleosidase activity / Toll Like Receptor 3 (TLR3) Cascade / NAD+ catabolic process / NAD+ nucleosidase activity / regulation of synapse pruning / modification of postsynaptic structure / ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase / protein localization to mitochondrion / NAD+ nucleosidase activity, cyclic ADP-ribose generating / nervous system process / Hydrolases; Glycosylases; Hydrolysing N-glycosyl compounds / regulation of dendrite morphogenesis / response to glucose / response to axon injury / signaling adaptor activity / TRAF6-mediated induction of TAK1 complex within TLR4 complex / regulation of neuron apoptotic process / Activation of IRF3, IRF7 mediated by TBK1, IKKε (IKBKE) / IKK complex recruitment mediated by RIP1 / neuromuscular junction / nervous system development / microtubule / mitochondrial outer membrane / cell differentiation / axon / innate immune response / dendrite / synapse / glutamatergic synapse / cell surface / signal transduction / protein-containing complex / mitochondrion / identical protein binding / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Cai Y / Zhang H | |||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: Permeant fluorescent probes visualize the activation of SARM1 and uncover an anti-neurodegenerative drug candidate. Authors: Wan Hua Li / Ke Huang / Yang Cai / Qian Wen Wang / Wen Jie Zhu / Yun Nan Hou / Sujing Wang / Sheng Cao / Zhi Ying Zhao / Xu Jie Xie / Yang Du / Chi-Sing Lee / Hon Cheung Lee / Hongmin Zhang / Yong Juan Zhao /  Abstract: SARM1 regulates axonal degeneration through its NAD-metabolizing activity and is a drug target for neurodegenerative disorders. We designed and synthesized fluorescent conjugates of styryl derivative ...SARM1 regulates axonal degeneration through its NAD-metabolizing activity and is a drug target for neurodegenerative disorders. We designed and synthesized fluorescent conjugates of styryl derivative with pyridine to serve as substrates of SARM1, which exhibited large red shifts after conversion. With the conjugates, SARM1 activation was visualized in live cells following elevation of endogenous NMN or treatment with a cell-permeant NMN-analog. In neurons, imaging documented mouse SARM1 activation preceded vincristine-induced axonal degeneration by hours. Library screening identified a derivative of nisoldipine (NSDP) as a covalent inhibitor of SARM1 that reacted with the cysteines, especially Cys311 in its ARM domain and blocked its NMN-activation, protecting axons from degeneration. The Cryo-EM structure showed that SARM1 was locked into an inactive conformation by the inhibitor, uncovering a potential neuroprotective mechanism of dihydropyridines. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30700.map.gz emd_30700.map.gz | 8.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30700-v30.xml emd-30700-v30.xml emd-30700.xml emd-30700.xml | 12.8 KB 12.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30700.png emd_30700.png | 168.1 KB | ||
| Filedesc metadata |  emd-30700.cif.gz emd-30700.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30700 http://ftp.pdbj.org/pub/emdb/structures/EMD-30700 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30700 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30700 | HTTPS FTP |
-Related structure data
| Related structure data |  7djtMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30700.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30700.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human SARM1 bounded with inhibitor dHNN | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.076 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
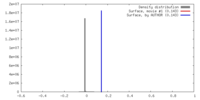
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human NAD(+) hydrolysis SARM1
| Entire | Name: Human NAD(+) hydrolysis SARM1 |
|---|---|
| Components |
|
-Supramolecule #1: Human NAD(+) hydrolysis SARM1
| Supramolecule | Name: Human NAD(+) hydrolysis SARM1 / type: cell / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: NAD(+) hydrolase SARM1
| Macromolecule | Name: NAD(+) hydrolase SARM1 / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO / EC number: ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 76.659656 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: RLAVPGPDGG GGTGPWWAAG GRGPREVSPG AGTEVQDALE RALPELQQAL SALKQAGGAR AVGAGLAEVF QLVEEAWLLP AVGREVAQG LCDAIRLDGG LDLLLRLLQA PELETRVQAA RLLEQILVAE NRDRVARIGL GVILNLAKER EPVELARSVA G ILEHMFKH ...String: RLAVPGPDGG GGTGPWWAAG GRGPREVSPG AGTEVQDALE RALPELQQAL SALKQAGGAR AVGAGLAEVF QLVEEAWLLP AVGREVAQG LCDAIRLDGG LDLLLRLLQA PELETRVQAA RLLEQILVAE NRDRVARIGL GVILNLAKER EPVELARSVA G ILEHMFKH SEETCQRLVA AGGLDAVLYW CRRTDPALLR HCALALGNCA LHGGQAVQRR MVEKRAAEWL FPLAFSKEDE LL RLHACLA VAVLATNKEV EREVERSGTL ALVEPLVASL DPGRFAR(VI3)LV DASDTSQGRG PDDLQRLVPL LDSNRLEAQ CIGAFYLCAE AAIKSLQGKT KVFSDIGAIQ SLKRLVSYST NGTKSALAKR ALRLLGEEVP RPILPSVPSW KEAEVQTWLQ QIGFSKYCE SFREQQVDGD LLLRLTEEEL QTDLGMKSGI TRKRFFRELT ELKTFANYST CDRSNLADWL GSLDPRFRQY T YGLVSCGL DRSLLHRVSE QQLLEDCGIH LGVHRARILT AAREMLHSPL PCTGGKPSGD TPDVFISYRR NSGSQLASLL KV HLQLHGF SVFIDVEKLE AGKFEDKLIQ SVMGARNFVL VLSPGALDKC MQDHDCKDWV HKEIVTALSC GKNIVPIIDG FEW PEPQVL PEDMQAVLTF NGIKWSHEYQ EATIEKIIRF LQGRSSRDSS AGSDTSLEGA APMGPT UniProtKB: NAD(+) hydrolase SARM1 |
-Macromolecule #2: O3-methyl O5-(2-methylpropyl) 2,6-dimethyl-4-[2-(oxidanylamino)ph...
| Macromolecule | Name: O3-methyl O5-(2-methylpropyl) 2,6-dimethyl-4-[2-(oxidanylamino)phenyl]pyridine-3,5-dicarboxylate type: ligand / ID: 2 / Number of copies: 8 / Formula: H8L |
|---|---|
| Molecular weight | Theoretical: 372.415 Da |
| Chemical component information |  ChemComp-H8L: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.00 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Details: BLOT TIME: 4S BLOT FORCE: -2. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Digitization - Frames/image: 4-36 / Number real images: 2673 / Average electron dose: 1.28 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: -2.5 µm / Calibrated defocus min: -2.0 µm / Calibrated magnification: 130000 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -0.8 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7djt: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)