[English] 日本語
 Yorodumi
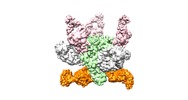
Yorodumi- EMDB-9189: Cryo-EM structure at 3.8 A resolution of HIV-1 fusion peptide-dir... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9189 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure at 3.8 A resolution of HIV-1 fusion peptide-directed antibody, DF1W-a.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / Fusion peptide / Neutralizing antibody / HIV-1 Envelope / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |    Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Acharya P / Xu K | |||||||||
 Citation Citation |  Journal: Cell / Year: 2019 Journal: Cell / Year: 2019Title: Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization. Authors: Rui Kong / Hongying Duan / Zizhang Sheng / Kai Xu / Priyamvada Acharya / Xuejun Chen / Cheng Cheng / Adam S Dingens / Jason Gorman / Mallika Sastry / Chen-Hsiang Shen / Baoshan Zhang / ...Authors: Rui Kong / Hongying Duan / Zizhang Sheng / Kai Xu / Priyamvada Acharya / Xuejun Chen / Cheng Cheng / Adam S Dingens / Jason Gorman / Mallika Sastry / Chen-Hsiang Shen / Baoshan Zhang / Tongqing Zhou / Gwo-Yu Chuang / Cara W Chao / Ying Gu / Alexander J Jafari / Mark K Louder / Sijy O'Dell / Ariana P Rowshan / Elise G Viox / Yiran Wang / Chang W Choi / Martin M Corcoran / Angela R Corrigan / Venkata P Dandey / Edward T Eng / Hui Geng / Kathryn E Foulds / Yicheng Guo / Young D Kwon / Bob Lin / Kevin Liu / Rosemarie D Mason / Martha C Nason / Tiffany Y Ohr / Li Ou / Reda Rawi / Edward K Sarfo / Arne Schön / John P Todd / Shuishu Wang / Hui Wei / Winston Wu / / James C Mullikin / Robert T Bailer / Nicole A Doria-Rose / Gunilla B Karlsson Hedestam / Diana G Scorpio / Julie Overbaugh / Jesse D Bloom / Bridget Carragher / Clinton S Potter / Lawrence Shapiro / Peter D Kwong / John R Mascola /   Abstract: The vaccine-mediated elicitation of antibodies (Abs) capable of neutralizing diverse HIV-1 strains has been a long-standing goal. To understand how broadly neutralizing antibodies (bNAbs) can be ...The vaccine-mediated elicitation of antibodies (Abs) capable of neutralizing diverse HIV-1 strains has been a long-standing goal. To understand how broadly neutralizing antibodies (bNAbs) can be elicited, we identified, characterized, and tracked five neutralizing Ab lineages targeting the HIV-1-fusion peptide (FP) in vaccinated macaques over time. Genetic and structural analyses revealed two of these lineages to belong to a reproducible class capable of neutralizing up to 59% of 208 diverse viral strains. B cell analysis indicated each of the five lineages to have been initiated and expanded by FP-carrier priming, with envelope (Env)-trimer boosts inducing cross-reactive neutralization. These Abs had binding-energy hotspots focused on FP, whereas several FP-directed Abs induced by immunization with Env trimer-only were less FP-focused and less broadly neutralizing. Priming with a conserved subregion, such as FP, can thus induce Abs with binding-energy hotspots coincident with the target subregion and capable of broad neutralization. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9189.map.gz emd_9189.map.gz | 14 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9189-v30.xml emd-9189-v30.xml emd-9189.xml emd-9189.xml | 23.6 KB 23.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9189.png emd_9189.png | 151.8 KB | ||
| Filedesc metadata |  emd-9189.cif.gz emd-9189.cif.gz | 7.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9189 http://ftp.pdbj.org/pub/emdb/structures/EMD-9189 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9189 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9189 | HTTPS FTP |
-Related structure data
| Related structure data |  6mphMC  8977C  9319C  9320C  9359C  6mpgC  6mqcC  6mqeC  6mqmC  6mqrC  6mqsC  6n16C  6n1vC  6n1wC  6nf2C  6osyC  6ot1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9189.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9189.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : DF1W-a.01 Fab bound to HIV-1 Env BG505 DS-SOSIP-VRC03 Fab- PGT122 Fab
+Supramolecule #1: DF1W-a.01 Fab bound to HIV-1 Env BG505 DS-SOSIP-VRC03 Fab- PGT122 Fab
+Supramolecule #2: DF1W-a.01
+Supramolecule #3: Envelope glycoprotein gp120
+Supramolecule #4: Envelope glycoprotein gp41
+Supramolecule #5: PGT122
+Supramolecule #6: VRC03
+Macromolecule #1: DF1W-a.01 heavy chain
+Macromolecule #2: DF1W-a.01 Light chain
+Macromolecule #3: Envelope glycoprotein gp41
+Macromolecule #4: Envelope glycoprotein gp120
+Macromolecule #5: PGT122 heavy chain
+Macromolecule #6: PGT122 Light Chain
+Macromolecule #7: VRC03 Heavy chain
+Macromolecule #8: VRC03 Light Chain
+Macromolecule #14: 2-acetamido-2-deoxy-beta-D-glucopyranose
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.25 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Instrument: SPOTITON |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.78 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















