[English] 日本語
 Yorodumi
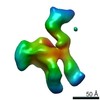
Yorodumi- EMDB-22443: Cryo-EM structure of ABCG5/G8 in complex with Fab 2E10 and 11F4 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22443 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of ABCG5/G8 in complex with Fab 2E10 and 11F4 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ABC transporter / Sterol / ATP hydrolysis / Pump / Fab / complex / Cholesterol / membrane protein / TRANSLOCASE-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of intestinal phytosterol absorption / negative regulation of intestinal cholesterol absorption / Defective ABCG8 causes GBD4 and sitosterolemia / Defective ABCG5 causes sitosterolemia / ABC transporters in lipid homeostasis / sterol transport / intestinal cholesterol absorption / phospholipid transport / cholesterol transfer activity / Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate ...negative regulation of intestinal phytosterol absorption / negative regulation of intestinal cholesterol absorption / Defective ABCG8 causes GBD4 and sitosterolemia / Defective ABCG5 causes sitosterolemia / ABC transporters in lipid homeostasis / sterol transport / intestinal cholesterol absorption / phospholipid transport / cholesterol transfer activity / Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate / cholesterol efflux / triglyceride homeostasis / response to muscle activity / response to ionizing radiation / ATPase-coupled transmembrane transporter activity / ABC-type transporter activity / NR1H3 & NR1H2 regulate gene expression linked to cholesterol transport and efflux / response to nutrient / ATP-binding cassette (ABC) transporter complex / cholesterol homeostasis / transmembrane transport / receptor complex / apical plasma membrane / response to xenobiotic stimulus / protein heterodimerization activity / ATP hydrolysis activity / ATP binding / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Huang CS / Yu X | |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2021 Journal: Commun Biol / Year: 2021Title: Cryo-EM structure of ABCG5/G8 in complex with modulating antibodies. Authors: Hanzhi Zhang / Ching-Shin Huang / Xinchao Yu / Jonas Lee / Amit Vaish / Qing Chen / Mingyue Zhou / Zhulun Wang / Xiaoshan Min /  Abstract: The heterodimer of ATP-binding cassette transporter ABCG5 and ABCG8 mediates the excretion of sterols from liver and intestine, playing a critical role in cholesterol homeostasis. Here, we present ...The heterodimer of ATP-binding cassette transporter ABCG5 and ABCG8 mediates the excretion of sterols from liver and intestine, playing a critical role in cholesterol homeostasis. Here, we present the cryo-EM structure of ABCG5/G8 in complex with the Fab fragments from two monoclonal antibodies at 3.3Å resolution. The high-resolution structure reveals a unique dimer interface between the nucleotide-binding domains (NBD) of opposing transporters, consisting of an ordered network of salt bridges between the conserved NPXDFXXD motif and serving as a pivot point that may be important for the transport cycle. While mAb 11F4 increases the ATPase activity potentially by stabilization of the NBD dimer formation, mAb 2E10 inhibits ATP hydrolysis, likely by restricting the relative movement between the RecA and helical domain of ABCG8 NBD. Our study not only provides insights into the structural elements important for the transport cycle but also reveals novel epitopes for potential therapeutic interventions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22443.map.gz emd_22443.map.gz | 166.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22443-v30.xml emd-22443-v30.xml emd-22443.xml emd-22443.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22443.png emd_22443.png | 69.4 KB | ||
| Filedesc metadata |  emd-22443.cif.gz emd-22443.cif.gz | 6.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22443 http://ftp.pdbj.org/pub/emdb/structures/EMD-22443 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22443 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22443 | HTTPS FTP |
-Related structure data
| Related structure data |  7jr7MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22443.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22443.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0588 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : ABCG5/G8 in complex with Fab 2E10 and 11F4
+Supramolecule #1: ABCG5/G8 in complex with Fab 2E10 and 11F4
+Supramolecule #2: ABCG5/G8
+Supramolecule #3: Fab 11F4
+Supramolecule #4: Fab 2E10
+Macromolecule #1: ATP-binding cassette sub-family G member 5
+Macromolecule #2: ATP-binding cassette sub-family G member 8
+Macromolecule #3: Fab 11F4 heavy chain
+Macromolecule #4: Fab 11F4 light chain
+Macromolecule #5: Fab 2E10 heavy chain
+Macromolecule #6: Fab 2E10 light chain
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 47.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.3 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 492931 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 3.0) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 3.0) |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















 Komagataella pastoris (fungus)
Komagataella pastoris (fungus)