-Search query
-Search result
Showing 1 - 50 of 250 items for (author: lander & g)

EMDB-43161: 
Human transthyretin covalently modified with A2-derived stilbene in the canonical conformation
Method: single particle / : Basanta B, Nugroho K, Yan N, Kline GM, Tsai FJ, Wu M, Kelly JW, Lander GC

EMDB-43162: 
Unliganded human transthyretin in the canonical conformation
Method: single particle / : Basanta B, Nugroho K, Yan N, Kline GM, Tsai FJ, Wu M, Kelly JW, Lander GC

EMDB-43163: 
Unliganded human transthyretin in the compressed conformation
Method: single particle / : Basanta B, Nugroho K, Yan N, Kline GM, Tsai FJ, Wu M, Kelly JW, Lander GC

EMDB-43164: 
Unliganded human transthyretin in the frayed conformation
Method: single particle / : Basanta B, Nugroho K, Yan N, Kline GM, Tsai FJ, Wu M, Kelly JW, Lander GC

EMDB-43165: 
(Biarylamine-FT2-WT)1(C10A)3-human transthyretin in the compressed conformation
Method: single particle / : Basanta B, Nugroho K, Yan N, Kline GM, Tsai FJ, Wu M, Kelly JW, Lander GC

EMDB-43166: 
(Biarylamine-FT2-WT)1(C10A)3-human transthyretin in the frayed conformation
Method: single particle / : Basanta B, Nugroho K, Yan N, Kline GM, Tsai FJ, Wu M, Kelly JW, Lander GC

PDB-8ve1: 
Human transthyretin covalently modified with A2-derived stilbene in the canonical conformation
Method: single particle / : Basanta B, Nugroho K, Yan N, Kline GM, Tsai FJ, Wu M, Kelly JW, Lander GC

PDB-8ve2: 
Unliganded human transthyretin in the canonical conformation
Method: single particle / : Basanta B, Nugroho K, Yan N, Kline GM, Tsai FJ, Wu M, Kelly JW, Lander GC

PDB-8ve3: 
Unliganded human transthyretin in the compressed conformation
Method: single particle / : Basanta B, Nugroho K, Yan N, Kline GM, Tsai FJ, Wu M, Kelly JW, Lander GC

PDB-8ve4: 
Unliganded human transthyretin in the frayed conformation
Method: single particle / : Basanta B, Nugroho K, Yan N, Kline GM, Tsai FJ, Wu M, Kelly JW, Lander GC

PDB-8ve5: 
(Biarylamine-FT2-WT)1(C10A)3-human transthyretin in the compressed conformation
Method: single particle / : Basanta B, Nugroho K, Yan N, Kline GM, Tsai FJ, Wu M, Kelly JW, Lander GC

PDB-8ve6: 
(Biarylamine-FT2-WT)1(C10A)3-human transthyretin in the frayed conformation
Method: single particle / : Basanta B, Nugroho K, Yan N, Kline GM, Tsai FJ, Wu M, Kelly JW, Lander GC

EMDB-42884: 
Microtubule-TTLL6 map
Method: single particle / : Mahalingan KK, Grotjahn D, Li Y, Lander GC, Zehr EA, Roll-Mecak A

PDB-8u3z: 
Model of TTLL6 bound to microtubule from composite map
Method: single particle / : Mahalingan KK, Grotjahn D, Li Y, Lander GC, Zehr EA, Roll-Mecak A

EMDB-41022: 
Map of Tubulin Tyrosine Ligase Like 6
Method: single particle / : Mahalingan KK, Grotjahn D, Li Y, Lander GC, Zehr EA, Roll-Mecak A
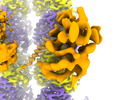
EMDB-41090: 
Composite map of TTLL6 bound to unmodified human microtubule
Method: single particle / : Mahalingan KK, Grotjahn D, Li Y, Lander GC, Zehr EA, Roll-Mecak A
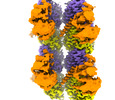
EMDB-41018: 
Microtubule-TTLL6 map
Method: single particle / : Mahalingan KK, Grotjahn D, Li Y, Lander GC, Zehr EA, Roll-Mecak A

PDB-8t42: 
Model of TTLL6 MTBH1-2 bound to microtubule
Method: single particle / : Mahalingan KK, Grotjahn D, Li Y, Lander GC, Zehr EA, Roll-Mecak A

EMDB-44534: 
Human DNA polymerase theta helicase domain tetramer in the apo form
Method: single particle / : Zerio CJ, Lander GC

EMDB-44535: 
Human DNA polymerase theta helicase domain dimer in the apo form
Method: single particle / : Zerio CJ, Lander GC

EMDB-44536: 
Human DNA polymerase theta helicase domain dimer bound to DNA in the microhomology searching conformation
Method: single particle / : Zerio CJ, Lander GC

EMDB-44537: 
Human DNA polymerase theta helicase domain dimer bound to DNA in the microhomology aligning conformation
Method: single particle / : Zerio CJ, Lander GC

EMDB-44538: 
Human DNA polymerase theta helicase domain dimer bound to DNA in the microhomology annealed conformation
Method: single particle / : Zerio CJ, Lander GC

PDB-9bh6: 
Human DNA polymerase theta helicase domain tetramer in the apo form
Method: single particle / : Zerio CJ, Lander GC

PDB-9bh7: 
Human DNA polymerase theta helicase domain dimer in the apo form
Method: single particle / : Zerio CJ, Lander GC

PDB-9bh8: 
Human DNA polymerase theta helicase domain dimer bound to DNA in the microhomology searching conformation
Method: single particle / : Zerio CJ, Lander GC

PDB-9bh9: 
Human DNA polymerase theta helicase domain dimer bound to DNA in the microhomology aligning conformation
Method: single particle / : Zerio CJ, Lander GC

PDB-9bha: 
Human DNA polymerase theta helicase domain dimer bound to DNA in the microhomology annealed conformation
Method: single particle / : Zerio CJ, Lander GC

EMDB-43160: 
Human transthyretin covalently modified with A2-derived stilbene in the compressed conformation
Method: single particle / : Basanta B, Nugroho K, Yan N, Kline GM, Tsai FJ, Wu M, Kelly JW, Lander GC

PDB-8ve0: 
Human transthyretin covalently modified with A2-derived stilbene in the compressed conformation
Method: single particle / : Basanta B, Nugroho K, Yan N, Kline GM, Tsai FJ, Wu M, Kelly JW, Lander GC

EMDB-41788: 
S. cerevisiae Pex1/Pex6 with 1 mM ATP
Method: single particle / : Gardner BM, Richardson CD, Martin A, Lander GC, Chowdhury S

PDB-8u0v: 
S. cerevisiae Pex1/Pex6 with 1 mM ATP
Method: single particle / : Gardner BM

EMDB-29695: 
CryoEM structure of yeast recombination mediator Rad52
Method: single particle / : Deveryshetty J, Basore K, Rau M, Fitzpatrick JAJ, Antony E

PDB-8g3g: 
CryoEM structure of yeast recombination mediator Rad52
Method: single particle / : Deveryshetty J, Basore K, Rau M, Fitzpatrick JAJ, Antony E

EMDB-29280: 
Cas1-Cas2/3 integrase and IHF bound to CRISPR leader, repeat and foreign DNA
Method: single particle / : Santiago-Frangos A, Henriques WS, Wiegand T, Gauvin C, Buyukyoruk M, Neselu K, Eng ET, Lander GC, Wiedenheft B

PDB-8flj: 
Cas1-Cas2/3 integrase and IHF bound to CRISPR leader, repeat and foreign DNA
Method: single particle / : Santiago-Frangos A, Henriques WS, Wiegand T, Gauvin C, Buyukyoruk M, Neselu K, Eng ET, Lander GC, Wiedenheft B

EMDB-29070: 
Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA(UGA-TL)
Method: single particle / : Hirschi M, Kuhle B, Doerfel L, Schimmel P, Lander G

PDB-8ffy: 
Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA(UGA-TL)
Method: single particle / : Hirschi M, Kuhle B, Doerfel L, Schimmel P, Lander G

EMDB-27269: 
Cryo-EM structure of human DELE1 in oligomeric form
Method: single particle / : Yang J, Lander GC

PDB-8d9x: 
Cryo-EM structure of human DELE1 in oligomeric form
Method: single particle / : Yang J, Lander GC
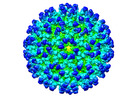
EMDB-26945: 
Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a moderately/weakly neutralizing human antibody IgG-21
Method: single particle / : Bandyopadhyay A, Klose T, Kuhn RJ
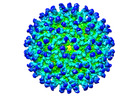
EMDB-26946: 
Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a moderately/weakly neutralizing human antibody IgG-94
Method: single particle / : Bandyopadhyay A, Klose T, Kuhn RJ
EMDB-26947: 
Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a potently neutralizing human antibody IgG-106
Method: single particle / : Bandyopadhyay A, Klose T, Kuhn RJ

PDB-7v0n: 
Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a moderately/weakly neutralizing human antibody IgG-21
Method: single particle / : Bandyopadhyay A, Klose T, Kuhn RJ

PDB-7v0o: 
Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a moderately/weakly neutralizing human antibody IgG-94
Method: single particle / : Bandyopadhyay A, Klose T, Kuhn RJ

PDB-7v0p: 
Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a potently neutralizing human antibody IgG-106
Method: single particle / : Bandyopadhyay A, Klose T, Kuhn RJ

EMDB-25730: 
Cryo-EM structure of full-length hepatitis C virus E1E2 glycoprotein in complex with AR4A, AT12009, and IGH505 Fabs
Method: single particle / : Torrents de la Pena A, Ward AB, Eshun-Wilson L, Lander GC

EMDB-27578: 
nsEM map of E1E2 AMS0232 glycoprotein in complex with monoclonal antibody AR4A
Method: single particle / : Torrents de la Pena A, Ward AB

PDB-7t6x: 
Cryo-EM structure of full-length hepatitis C virus E1E2 glycoprotein in complex with AR4A, AT12009, and IGH505 Fabs
Method: single particle / : Torrents de la Pena A, Ward AB, Eshun-Wilson L, Lander GC
EMDB-26310: 
Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA (GCU)
Method: single particle / : Hirschi M, Kuhle B, Doerfel L, Schimmel P, Lander G
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
