[English] 日本語
 Yorodumi
Yorodumi- EMDB-25730: Cryo-EM structure of full-length hepatitis C virus E1E2 glycoprot... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of full-length hepatitis C virus E1E2 glycoprotein in complex with AR4A, AT12009, and IGH505 Fabs | |||||||||
 Map data Map data | Sharpened non-focused map of the full complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | hepatitis C virus / E1E2 / viral protein-immune system complex / antibody-antigen complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell mitochondrial membrane / host cell lipid droplet / viral nucleocapsid / clathrin-dependent endocytosis of virus by host cell / symbiont-mediated suppression of host innate immune response / host cell endoplasmic reticulum membrane / ribonucleoprotein complex / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...host cell mitochondrial membrane / host cell lipid droplet / viral nucleocapsid / clathrin-dependent endocytosis of virus by host cell / symbiont-mediated suppression of host innate immune response / host cell endoplasmic reticulum membrane / ribonucleoprotein complex / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell nucleus / virion membrane / structural molecule activity / RNA binding Similarity search - Function | |||||||||
| Biological species |  Hepacivirus C / Hepacivirus C /  Homo sapiens (human) Homo sapiens (human) | |||||||||
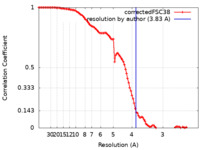
| Method | single particle reconstruction / cryo EM / Resolution: 3.83 Å | |||||||||
 Authors Authors | Torrents de la Pena A / Ward AB / Eshun-Wilson L / Lander GC | |||||||||
| Funding support |  Netherlands, 1 items Netherlands, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Structure of the hepatitis C virus E1E2 glycoprotein complex. Authors: Alba Torrents de la Peña / Kwinten Sliepen / Lisa Eshun-Wilson / Maddy L Newby / Joel D Allen / Ian Zon / Sylvie Koekkoek / Ana Chumbe / Max Crispin / Janke Schinkel / Gabriel C Lander / ...Authors: Alba Torrents de la Peña / Kwinten Sliepen / Lisa Eshun-Wilson / Maddy L Newby / Joel D Allen / Ian Zon / Sylvie Koekkoek / Ana Chumbe / Max Crispin / Janke Schinkel / Gabriel C Lander / Rogier W Sanders / Andrew B Ward /    Abstract: Hepatitis C virus (HCV) infection is a leading cause of chronic liver disease, cirrhosis, and hepatocellular carcinoma in humans and afflicts more than 58 million people worldwide. The HCV envelope ...Hepatitis C virus (HCV) infection is a leading cause of chronic liver disease, cirrhosis, and hepatocellular carcinoma in humans and afflicts more than 58 million people worldwide. The HCV envelope E1 and E2 glycoproteins are essential for viral entry and comprise the primary antigenic target for neutralizing antibody responses. The molecular mechanisms of E1E2 assembly, as well as how the E1E2 heterodimer binds broadly neutralizing antibodies, remain elusive. Here, we present the cryo-electron microscopy structure of the membrane-extracted full-length E1E2 heterodimer in complex with three broadly neutralizing antibodies-AR4A, AT1209, and IGH505-at ~3.5-angstrom resolution. We resolve the interface between the E1 and E2 ectodomains and deliver a blueprint for the rational design of vaccine immunogens and antiviral drugs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25730.map.gz emd_25730.map.gz | 97.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25730-v30.xml emd-25730-v30.xml emd-25730.xml emd-25730.xml | 31.5 KB 31.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25730_fsc.xml emd_25730_fsc.xml | 10.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_25730.png emd_25730.png | 36.7 KB | ||
| Masks |  emd_25730_msk_1.map emd_25730_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-25730.cif.gz emd-25730.cif.gz | 8.1 KB | ||
| Others |  emd_25730_additional_1.map.gz emd_25730_additional_1.map.gz emd_25730_additional_2.map.gz emd_25730_additional_2.map.gz emd_25730_half_map_1.map.gz emd_25730_half_map_1.map.gz emd_25730_half_map_2.map.gz emd_25730_half_map_2.map.gz | 97.2 MB 51.8 MB 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25730 http://ftp.pdbj.org/pub/emdb/structures/EMD-25730 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25730 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25730 | HTTPS FTP |
-Related structure data
| Related structure data |  7t6xMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25730.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25730.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened non-focused map of the full complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||
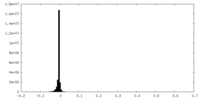
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_25730_msk_1.map emd_25730_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
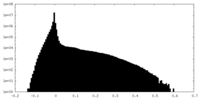
| Density Histograms |
-Additional map: Composite sharpened map of the complex generated from...
| File | emd_25730_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite sharpened map of the complex generated from multiple focused maps of different sub-regions of the complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
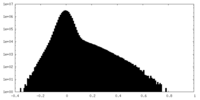
| Density Histograms |
-Additional map: Unsharpened non-focused map of the full complex
| File | emd_25730_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened non-focused map of the full complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
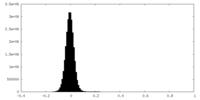
| Density Histograms |
-Half map: Half map of sharpened non-focused map of full complex
| File | emd_25730_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of sharpened non-focused map of full complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map of sharpened non-focused map of full complex
| File | emd_25730_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of sharpened non-focused map of full complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : cryoEM structure of full-length hepatitis C virus E1E2 glycoprote...
+Supramolecule #1: cryoEM structure of full-length hepatitis C virus E1E2 glycoprote...
+Macromolecule #1: Envelope glycoprotein E2
+Macromolecule #2: AR4A Fab, heavy chain
+Macromolecule #3: AR4A, kappa light chain
+Macromolecule #4: IGH505, heavy chain
+Macromolecule #5: IGH505, lambda light chain
+Macromolecule #6: AT12009 crossMab, light kappa chain
+Macromolecule #7: AT12009 CrossMab, heavy chain
+Macromolecule #8: Envelope glycoprotein E1
+Macromolecule #13: 2-acetamido-2-deoxy-beta-D-glucopyranose
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: TBS buffer, pH 7.4 | |||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Support film - Film thickness: 50 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 7 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 40 K / Instrument: FEI VITROBOT MARK IV Details: Blotting time: 5 s; Blotting force: 0; Waiting time: 7 s. | |||||||||
| Details | Full-length hepatitis C virus E1E2 glycoprotein complexed with AR4A, AT12009, and IGH505 Fabs and embedded in peptidiscs is diluted in TBS to a final concentration of 0.8 mg/ml. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 2 / Number real images: 5121 / Average exposure time: 9.0 sec. / Average electron dose: 50.0 e/Å2 Details: images were collected at 250 ms per frame. 36 frames were collected. |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 43478 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.12 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 36000 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)