[English] 日本語
 Yorodumi
Yorodumi- EMDB-29070: Cryo-electron microscopy structure of human mt-SerRS in complex w... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA(UGA-TL) | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | tRNA / SerRS / transcription / ligase-RNA complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationmitochondrial seryl-tRNA aminoacylation / Mitochondrial tRNA aminoacylation / serine-tRNA ligase / serine-tRNA ligase activity / seryl-tRNA aminoacylation / tRNA binding / mitochondrial matrix / mitochondrion / RNA binding / ATP binding / cytosol Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||||||||
 Authors Authors | Hirschi M / Kuhle B / Doerfel L / Schimmel P / Lander G | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis for a degenerate tRNA identity code and the evolution of bimodal specificity in human mitochondrial tRNA recognition. Authors: Bernhard Kuhle / Marscha Hirschi / Lili K Doerfel / Gabriel C Lander / Paul Schimmel /   Abstract: Animal mitochondrial gene expression relies on specific interactions between nuclear-encoded aminoacyl-tRNA synthetases and mitochondria-encoded tRNAs. Their evolution involves an antagonistic ...Animal mitochondrial gene expression relies on specific interactions between nuclear-encoded aminoacyl-tRNA synthetases and mitochondria-encoded tRNAs. Their evolution involves an antagonistic interplay between strong mutation pressure on mtRNAs and selection pressure to maintain their essential function. To understand the molecular consequences of this interplay, we analyze the human mitochondrial serylation system, in which one synthetase charges two highly divergent mtRNA isoacceptors. We present the cryo-EM structure of human mSerRS in complex with mtRNA, and perform a structural and functional comparison with the mSerRS-mtRNA complex. We find that despite their common function, mtRNA and mtRNA show no constrain to converge on shared structural or sequence identity motifs for recognition by mSerRS. Instead, mSerRS evolved a bimodal readout mechanism, whereby a single protein surface recognizes degenerate identity features specific to each mtRNA. Our results show how the mutational erosion of mtRNAs drove a remarkable innovation of intermolecular specificity rules, with multiple evolutionary pathways leading to functionally equivalent outcomes. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29070.map.gz emd_29070.map.gz | 25.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29070-v30.xml emd-29070-v30.xml emd-29070.xml emd-29070.xml | 17 KB 17 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29070.png emd_29070.png | 112.9 KB | ||
| Others |  emd_29070_half_map_1.map.gz emd_29070_half_map_1.map.gz emd_29070_half_map_2.map.gz emd_29070_half_map_2.map.gz | 20.6 MB 20.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29070 http://ftp.pdbj.org/pub/emdb/structures/EMD-29070 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29070 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29070 | HTTPS FTP |
-Related structure data
| Related structure data |  8ffyMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29070.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29070.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_29070_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
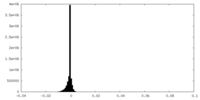
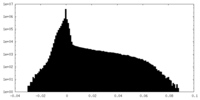
| Density Histograms |
-Half map: #1
| File | emd_29070_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
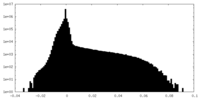
| Density Histograms |
- Sample components
Sample components
-Entire : Human mt-SerRS in complex with mt-tRNA(UGA-TL) and SerSA
| Entire | Name: Human mt-SerRS in complex with mt-tRNA(UGA-TL) and SerSA |
|---|---|
| Components |
|
-Supramolecule #1: Human mt-SerRS in complex with mt-tRNA(UGA-TL) and SerSA
| Supramolecule | Name: Human mt-SerRS in complex with mt-tRNA(UGA-TL) and SerSA type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Serine--tRNA ligase, mitochondrial
| Macromolecule | Name: Serine--tRNA ligase, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: serine-tRNA ligase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 58.358391 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAASMARRLW PLLTRRGFRP RGGCISNDSP RRSFTTEKRN RNLLYEYARE GYSALPQLDI ERFCACPEEA AHALELRKGE LRSADLPAI ISTWQELRQL QEQIRSLEEE KAAVTEAVRA LLANQDSGEV QQDPKYQGLR ARGREIRKEL VHLYPREAQL E EQFYLQAL ...String: MAASMARRLW PLLTRRGFRP RGGCISNDSP RRSFTTEKRN RNLLYEYARE GYSALPQLDI ERFCACPEEA AHALELRKGE LRSADLPAI ISTWQELRQL QEQIRSLEEE KAAVTEAVRA LLANQDSGEV QQDPKYQGLR ARGREIRKEL VHLYPREAQL E EQFYLQAL KLPNQTHPDV PVGDESQARV LHMVGDKPVF SFQPRGHLEI GEKLDIIRQK RLSHVSGHRS YYLRGAGALL QH GLVNFTF NKLLRRGFTP MTVPDLLRGA VFEGCGMTPN ANPSQIYNID PARFKDLNLA GTAEVGLAGY FMDHTVAFRD LPV RMVCSS TCYRAETNTG QEPRGLYRVH HFTKVEMFGV TGPGLEQSSQ LLEEFLSLQM EILTELGLHF RVLDMPTQEL GLPA YRKFD IEAWMPGRGR FGEVTSASNC TDFQSRRLHI MFQTEAGELQ FAHTVNATAC AVPRLLIALL ESNQQKDGSV LVPPA LQSY LGTDRITAPT HVPLQYIGPN QPRKPGLPGQ PAVS UniProtKB: Serine--tRNA ligase, mitochondrial |
-Macromolecule #2: mt-tRNA(UGA-TL)
| Macromolecule | Name: mt-tRNA(UGA-TL) / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 21.41141 KDa |
| Sequence | String: GAAAAAGUCA UGGAGGCCAU GGGGUCC(N)(N)(N) (N)(N)(N)(N)(N)(N)GGGC UUUGGGGGGU UCGAUUCCUU CC UUUUUUG C |
-Macromolecule #3: 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE
| Macromolecule | Name: 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE / type: ligand / ID: 3 / Number of copies: 2 / Formula: SSA |
|---|---|
| Molecular weight | Theoretical: 433.397 Da |
| Chemical component information | 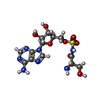 ChemComp-SSA: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 7 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 3448 / Average exposure time: 11.8 sec. / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8ffy: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)