1A85
 
 | | MMP8 WITH MALONIC AND ASPARAGINE BASED INHIBITOR | | Descriptor: | CALCIUM ION, MMP-8, N~1~-(3-aminobenzyl)-N~2~-[(2R)-2-(hydroxycarbamoyl)-4-methylpentanoyl]-L-aspartamide, ... | | Authors: | Brandstetter, H, Roedern, E.G.V, Grams, F, Engh, R.A. | | Deposit date: | 1998-04-03 | | Release date: | 1999-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of malonic acid-based inhibitors bound to human neutrophil collagenase. A new binding mode explains apparently anomalous data.
Protein Sci., 7, 1998
|
|
1A86
 
 | | MMP8 WITH MALONIC AND ASPARTIC ACID BASED INHIBITOR | | Descriptor: | CALCIUM ION, MMP-8, N-benzyl-N~2~-[(2R)-2-(hydroxycarbamoyl)-4-methylpentanoyl]-L-alpha-asparagine, ... | | Authors: | Brandstetter, H, Roedern, E.G.V, Grams, F, Engh, R.A. | | Deposit date: | 1998-04-03 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of malonic acid-based inhibitors bound to human neutrophil collagenase. A new binding mode explains apparently anomalous data.
Protein Sci., 7, 1998
|
|
1FAX
 
 | | COAGULATION FACTOR XA INHIBITOR COMPLEX | | Descriptor: | (2S)-3-(7-carbamimidoylnaphthalen-2-yl)-2-[4-({(3R)-1-[(1Z)-ethanimidoyl]pyrrolidin-3-yl}oxy)phenyl]propanoic acid, CALCIUM ION, FACTOR XA | | Authors: | Brandstetter, H, Engh, R.A. | | Deposit date: | 1996-08-23 | | Release date: | 1997-10-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of active site-inhibited clotting factor Xa. Implications for drug design and substrate recognition.
J.Biol.Chem., 271, 1996
|
|
1PFX
 
 | | PORCINE FACTOR IXA | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, FACTOR IXA | | Authors: | Brandstetter, H, Bauer, M, Huber, R, Lollar, P, Bode, W. | | Deposit date: | 1995-07-19 | | Release date: | 1996-08-17 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of clotting factor IXa: active site and module structure related to Xase activity and hemophilia B.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1JJ9
 
 | | Crystal Structure of MMP8-Barbiturate Complex Reveals Mechanism for Collagen Substrate Recognition | | Descriptor: | 2-HYDROXY-5-[4-(2-HYDROXY-ETHYL)-PIPERIDIN-1-YL]-5-PHENYL-1H-PYRIMIDINE-4,6-DIONE, CALCIUM ION, Matrix Metalloproteinase 8, ... | | Authors: | Brandstetter, H, Grams, F, Glitz, D, Lang, A, Huber, R, Bode, W, Krell, H.-W, Engh, R.A. | | Deposit date: | 2001-07-04 | | Release date: | 2001-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 1.8-A crystal structure of a matrix metalloproteinase 8-barbiturate inhibitor complex reveals a previously unobserved mechanism for collagenase substrate recognition.
J.Biol.Chem., 276, 2001
|
|
1K32
 
 | |
1ETT
 
 | |
1ETS
 
 | |
6FRR
 
 | | Structural and immunological properties of the allergen Art v 3 | | Descriptor: | Non-specific lipid-transfer protein, SULFATE ION | | Authors: | Brandstetter, H, Soh, W.T, Magler, I. | | Deposit date: | 2018-02-16 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Boiling down the cysteine-stabilized LTP fold - loss of structural and immunological integrity of allergenic Art v 3 and Pru p 3 as a consequence of irreversible lanthionine formation.
Mol.Immunol., 116, 2019
|
|
1ETR
 
 | | REFINED 2.3 ANGSTROMS X-RAY CRYSTAL STRUCTURE OF BOVINE THROMBIN COMPLEXES FORMED WITH THE BENZAMIDINE AND ARGININE-BASED THROMBIN INHIBITORS NAPAP, 4-TAPAP AND MQPA: A STARTING POINT FOR IMPROVING ANTITHROMBOTICS | | Descriptor: | EPSILON-THROMBIN, amino{[(4S)-5-[(2R,4R)-2-carboxy-4-methylpiperidin-1-yl]-4-({[(3R)-3-methyl-1,2,3,4-tetrahydroquinolin-8-yl]sulfonyl}amino)-5-oxopentyl]amino}methaniminium | | Authors: | Bode, W, Brandstetter, H. | | Deposit date: | 1992-07-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined 2.3 A X-ray crystal structure of bovine thrombin complexes formed with the benzamidine and arginine-based thrombin inhibitors NAPAP, 4-TAPAP and MQPA. A starting point for improving antithrombotics.
J.Mol.Biol., 226, 1992
|
|
6TRK
 
 | | Phl p 6 fold stabilized mutant - S46Y | | Descriptor: | Pollen allergen Phl p 6, ZINC ION | | Authors: | Soh, W.T, Brandstetter, H. | | Deposit date: | 2019-12-19 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In silico Design of Phl p 6 Variants With Altered Fold-Stability Significantly Impacts Antigen Processing, Immunogenicity and Immune Polarization.
Front Immunol, 11, 2020
|
|
5NZB
 
 | |
5OBT
 
 | |
1N6D
 
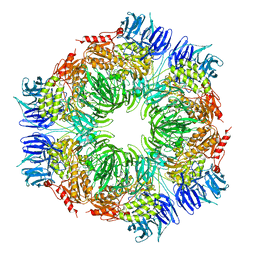 | | Tricorn protease in complex with tetrapeptide chloromethyl ketone derivative | | Descriptor: | DECANE, RVRK, Tricorn protease | | Authors: | Kim, J.-S, Groll, M, Huber, R, Brandstetter, H. | | Deposit date: | 2002-11-10 | | Release date: | 2002-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Navigation Inside a Protease: Substrate Selection and Product Exit in the Tricorn Protease
from Thermoplasma acidophilum
J.Mol.Biol., 324, 2002
|
|
7ZOC
 
 | |
6O2X
 
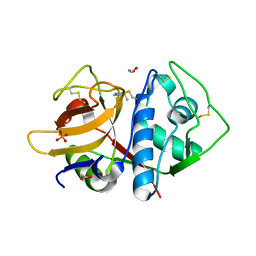 | | Structure of cruzain bound to MMTS inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Cruzipain, PHOSPHATE ION | | Authors: | Silva, E.B, Dall, E, Ferreira, R.S, Brandstetter, H. | | Deposit date: | 2019-02-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.193 Å) | | Cite: | Cruzain structures: apocruzain and cruzain bound to S-methyl thiomethanesulfonate and implications for drug design.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4QIP
 
 | |
6FOE
 
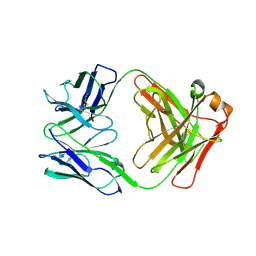 | | BaxB01 Fab fragment | | Descriptor: | Fab BaxB01 heavy chain, Fab BaxB01 light chain | | Authors: | Hollerweger, J, Schinagl, A, Kerschbaumer, R.J, Scheiflinger, F, Thiele, M, Goettig, P, Brandstetter, H. | | Deposit date: | 2018-02-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Role of the Cysteine 81 Residue of Macrophage Migration Inhibitory Factor as a Molecular Redox Switch.
Biochemistry, 57, 2018
|
|
6FK0
 
 | |
8OYH
 
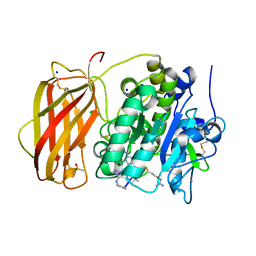 | | X-ray structure of furin (PCSK3) in complex with Guanidinomethyl-Phac-Can-Tle-Can-6-(aminomethyl)-3-amino-isoindol | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Dahms, S.O, Brandstetter, H. | | Deposit date: | 2023-05-04 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-Based Design, Synthesis, and Characterization of Aminoisoindole-Derived Furin Inhibitors.
Chemmedchem, 19, 2024
|
|
4BQD
 
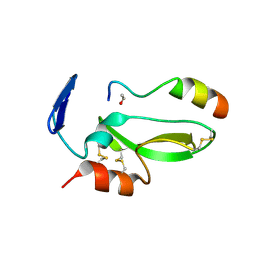 | | KD1 of human TFPI in complex with a synthetic peptide | | Descriptor: | GLYCEROL, PEPTIDE, TISSUE FACTOR PATHWAY INHIBITOR (LIPOPROTEIN-ASSOCIATED COAGULATION INHIBITOR) VARIANT | | Authors: | Griessner, A, Brandstetter, H. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-04 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Small Peptides Blocking Inhibition of Factor Xa and Tissue Factor-Factor Viia by Tissue Factor Pathway Inhibitor (Tfpi)
J.Biol.Chem., 289, 2014
|
|
2Y6I
 
 | |
7ZBV
 
 | |
7Z5U
 
 | | Crystal structure of the peptidase domain of collagenase G from Clostridium histolyticum in complex with a hydroxamate-based inhibitor | | Descriptor: | (2~{R})-~{N}-[2-[4-[(2-acetamidophenoxy)methyl]-1,2,3-triazol-1-yl]ethyl]-2-(2-methylpropyl)-~{N}'-oxidanyl-propanediamide, CALCIUM ION, Collagenase ColG, ... | | Authors: | Schoenauer, E, Brandstetter, H. | | Deposit date: | 2022-03-10 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and Characterization of Synthesized and FDA-Approved Inhibitors of Clostridial and Bacillary Collagenases.
J.Med.Chem., 65, 2022
|
|
6N3S
 
 | | Crystal structure of apo-cruzain | | Descriptor: | 1,2-ETHANEDIOL, Cruzipain, PHOSPHATE ION | | Authors: | Silva, E.B, Dall, E, Rodrigues, F.T.G, Ferreira, R.S, Brandstetter, H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.193 Å) | | Cite: | Cruzain structures: apocruzain and cruzain bound to S-methyl thiomethanesulfonate and implications for drug design.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
