7AUW
 
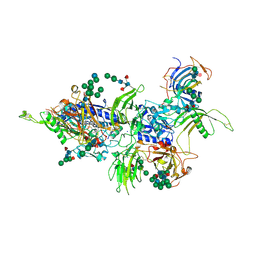 | | Inhibitory complex of human meprin beta with mouse fetuin-B. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Fetuin-B, ... | | Authors: | Eckhard, U, Gomis-Ruth, F.X. | | Deposit date: | 2020-11-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of a 250-kDa heterotetrameric particle explains inhibition of sheddase meprin beta by endogenous fetuin-B.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2Y72
 
 | |
7POQ
 
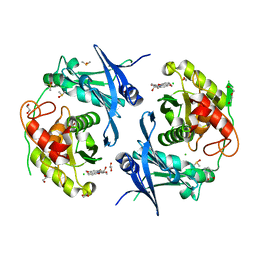 | | Crystal structure of profragilysin-3 (proBFT-3) from Bacteroides fragilis in complex with foliosidine in P41212. | | Descriptor: | 1,2-ETHANEDIOL, BFT-3, CHLORIDE ION, ... | | Authors: | Eckhard, U, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Repositioning small molecule drugs as allosteric inhibitors of the BFT-3 toxin from enterotoxigenic Bacteroides fragilis.
Protein Sci., 31, 2022
|
|
7PND
 
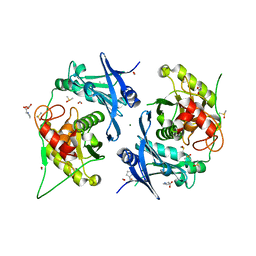 | | Crystal structure of profragilysin-3 (proBFT-3) from Bacteroides fragilis at 1.85 A resolution. | | Descriptor: | BFT-3, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Eckhard, U, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2021-09-06 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Repositioning small molecule drugs as allosteric inhibitors of the BFT-3 toxin from enterotoxigenic Bacteroides fragilis.
Protein Sci., 31, 2022
|
|
7POL
 
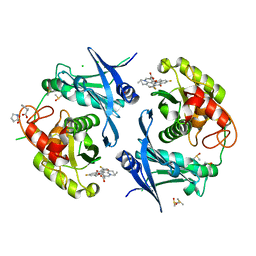 | | Crystal structure of profragilysin-3 (proBFT-3) from Bacteroides fragilis in complex with flumequine | | Descriptor: | (12~{R})-7-fluoranyl-12-methyl-4-oxidanylidene-1-azatricyclo[7.3.1.0^{5,13}]trideca-2,5(13),6,8-tetraene-3-carboxylic acid, BFT-3, CHLORIDE ION, ... | | Authors: | Eckhard, U, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Repositioning small molecule drugs as allosteric inhibitors of the BFT-3 toxin from enterotoxigenic Bacteroides fragilis.
Protein Sci., 31, 2022
|
|
2Y50
 
 | |
2Y6I
 
 | |
7POU
 
 | | Crystal structure of profragilysin-3 (proBFT-3) from Bacteroides fragilis in complex with hesperetin. | | Descriptor: | (2S)-5,7-dihydroxy-2-(3-hydroxy-4-methoxyphenyl)-2,3-dihydro-4H-1-benzopyran-4-one, BFT-3, DIMETHYL SULFOXIDE, ... | | Authors: | Eckhard, U, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Repositioning small molecule drugs as allosteric inhibitors of the BFT-3 toxin from enterotoxigenic Bacteroides fragilis.
Protein Sci., 31, 2022
|
|
7POO
 
 | | Crystal structure of profragilysin-3 (proBFT-3) from Bacteroides fragilis in complex with foliosidine in P212121. | | Descriptor: | ACETATE ION, BFT-3, PROLINE, ... | | Authors: | Eckhard, U, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Repositioning small molecule drugs as allosteric inhibitors of the BFT-3 toxin from enterotoxigenic Bacteroides fragilis.
Protein Sci., 31, 2022
|
|
2Y3U
 
 | |
6ZNB
 
 | | PHAGE SAM LYASE IN APO STATE | | Descriptor: | PHOSPHATE ION, Phage SAM lyase Svi3-3 | | Authors: | Eckhard, U, Kanchugal P, S, Selmer, M. | | Deposit date: | 2020-07-06 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanism of a phage-encoded SAM lyase revises catalytic function of enzyme family.
Elife, 10, 2021
|
|
4ARE
 
 | | Crystal structure of the collagenase Unit of collagenase G from Clostridium histolyticum at 2.19 angstrom resolution. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRATE ANION, COLLAGENASE G, ... | | Authors: | Eckhard, U, Brandstetter, H. | | Deposit date: | 2012-04-23 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Basis for Activity Regulation and Substrate Preference of Clostridial Collagenases G, H, and T.
J.Biol.Chem., 288, 2013
|
|
4ARF
 
 | | CRYSTAL STRUCTURE OF THE PEPTIDASE DOMAIN OF COLLAGENASE H FROM CLOSTRIDIUM HISTOLYTICUM IN COMPLEX WITH THE PEPTIDIC INHIBITOR ISOAMYLPHOSPHONYL-GLY-PRO-ALA AT 1.77 ANGSTROM RESOLUTION. | | Descriptor: | CALCIUM ION, COLH PROTEIN, ISOAMYLPHOSPHONYL-GLY-PRO-ALA, ... | | Authors: | Eckhard, U, Brandstetter, H. | | Deposit date: | 2012-04-23 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Basis for Activity Regulation and Substrate Preference of Clostridial Collagenases G, H, and T.
J.Biol.Chem., 288, 2013
|
|
4AR8
 
 | | Crystal structure of the peptidase domain of collagenase T from Clostridium tetani complexed with the peptidic inhibitor isoamyl- phosphonyl-Gly-Pro-Ala at 2.05 angstrom resolution. | | Descriptor: | CALCIUM ION, COLLAGENASE COLT, ISOAMYL-PHOSPHONYL-GLY-PRO-ALA, ... | | Authors: | Eckhard, U, Brandstetter, H. | | Deposit date: | 2012-04-21 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for Activity Regulation and Substrate Preference of Clostridial Collagenases G, H, and T.
J.Biol.Chem., 288, 2013
|
|
4AQO
 
 | |
4AR1
 
 | |
4AR9
 
 | |
8QB1
 
 | | C-terminal domain of mirolase from Tannerella forsythia | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Mirolase, ... | | Authors: | Gomis-Ruth, F.X, Rodriguez-Banqueri, A, Mizgalska, D, Veillard, F, Goulas, T, Eckhard, U, Potempa, J. | | Deposit date: | 2023-08-23 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and functional insights into the C-terminal signal domain of the Bacteroidetes type-IX secretion system
Open Biology, 2024
|
|
7SKM
 
 | | Complex between S. aureus aureolysin and wt IMPI. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mendes, S.R, Eckhard, U, Rodriguez-Banqueri, A, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2021-10-21 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An engineered protein-based submicromolar competitive inhibitor of the Staphylococcus aureus virulence factor aureolysin
Comput Struct Biotechnol J, 20, 2022
|
|
7SKN
 
 | |
7SKO
 
 | | De novo synthetic protein DIG8-CC (orthogonal space group) | | Descriptor: | De novo synthetic protein DIG8-CC, MAGNESIUM ION | | Authors: | Mendes, S.R, Eckhard, U, Marcos, E, Gomis-Ruth, F.X. | | Deposit date: | 2021-10-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | De novo design of immunoglobulin-like domains
Nat Commun, 13, 2022
|
|
7SKP
 
 | |
7SKL
 
 | | Complex between S. aureus aureolysin and IMPI mutant I57I | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, IMPI alpha, ... | | Authors: | Mendes, S.R, Eckhard, U, Rodriguez-Banqueri, A, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2021-10-21 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An engineered protein-based submicromolar competitive inhibitor of the Staphylococcus aureus virulence factor aureolysin
Comput Struct Biotechnol J, 20, 2022
|
|
8CDB
 
 | | Proulilysin E229A structure | | Descriptor: | CALCIUM ION, Ulilysin, ZINC ION | | Authors: | Rodriguez-Banqueri, A, Eckhard, U, Gomis-Ruth, F.X. | | Deposit date: | 2023-01-30 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural insights into latency of the metallopeptidase ulilysin (lysargiNase) and its unexpected inhibition by a sulfonyl-fluoride inhibitor of serine peptidases.
Dalton Trans, 52, 2023
|
|
8CD8
 
 | | Ulilysin - C269A with AEBSF complex | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CALCIUM ION, GLY-SER-SER, ... | | Authors: | Rodriguez-Banqueri, A, Eckhard, U, Gomis-Ruth, F.X. | | Deposit date: | 2023-01-30 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into latency of the metallopeptidase ulilysin (lysargiNase) and its unexpected inhibition by a sulfonyl-fluoride inhibitor of serine peptidases.
Dalton Trans, 52, 2023
|
|
