8PPL
 
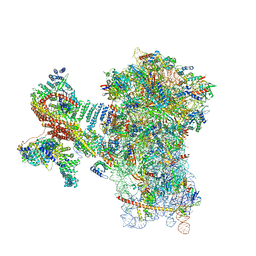 | | MERS-CoV Nsp1 bound to the human 43S pre-initiation complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | Deposit date: | 2023-07-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
8PPK
 
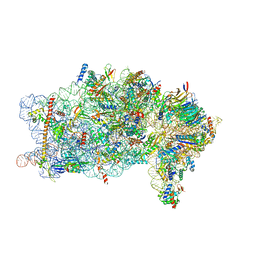 | | Bat-Hp-CoV Nsp1 and eIF1 bound to the human 40S small ribosomal subunit | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | Deposit date: | 2023-07-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
6ZOK
 
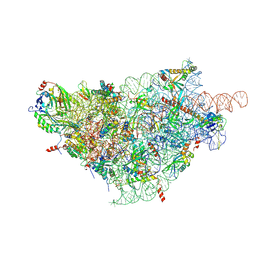 | | SARS-CoV-2-Nsp1-40S complex, focused on body | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Schubert, K, Karousis, E.D, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, L.-A, Leibundgut, M, Thiel, V, Muehlemann, O, Ban, N. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-29 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZOJ
 
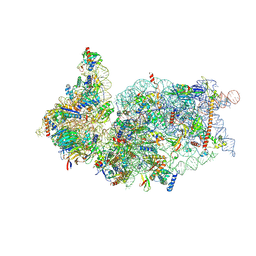 | | SARS-CoV-2-Nsp1-40S complex, composite map | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, L.-A, Leibundgut, M.L, Thiel, V, Muehlemann, O, Ban, N. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-22 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZOL
 
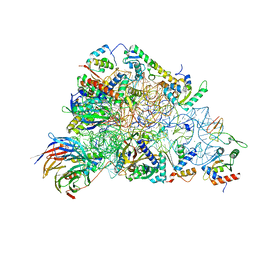 | | SARS-CoV-2-Nsp1-40S complex, focused on head | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S12, ... | | Authors: | Schubert, K, Karousis, E.D, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, L.-A, Leibundgut, M.L, Thiel, V, Muehlemann, O, Ban, N. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5LF8
 
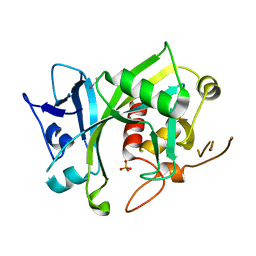 | | Human Nucleoside diphosphate-linked moiety X motif 17 (NUDT17) | | Descriptor: | ETHYL MERCURY ION, Nucleoside diphosphate-linked moiety X motif 17, PHOSPHATE ION | | Authors: | Mathea, S, Tallant, C, Salah, E, Wang, D, Velupillai, S, Nowak, R, Oerum, S, Krojer, T, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-30 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Human Nucleoside diphosphate-linked moiety X motif 17 (NUDT17)
To Be Published
|
|
3CXW
 
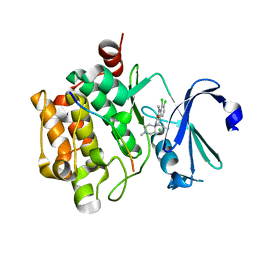 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a beta carboline ligand I | | Descriptor: | (4R)-7,8-dichloro-1',9-dimethyl-1-oxo-1,2,4,9-tetrahydrospiro[beta-carboline-3,4'-piperidine]-4-carbonitrile, CHLORIDE ION, Pimtide peptide, ... | | Authors: | Filippakopoulos, P, Bullock, A, Fedorov, O, Huber, K, Bracher, F, Pike, A.C.W, Roos, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 7,8-Dichloro-1-oxo-beta-carbolines as a Versatile Scaffold for the Development of Potent and Selective Kinase Inhibitors with Unusual Binding Modes
J.Med.Chem., 55, 2012
|
|
3CY2
 
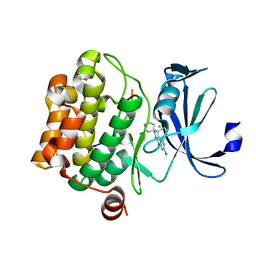 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a beta carboline ligand II | | Descriptor: | (4R)-7-chloro-9-methyl-1-oxo-1,2,4,9-tetrahydrospiro[beta-carboline-3,4'-piperidine]-4-carbonitrile, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Filippakopoulos, P, Bullock, A, Fedorov, O, Huber, K, Bracher, F, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | 7,8-Dichloro-1-oxo-beta-carbolines as a Versatile Scaffold for the Development of Potent and Selective Kinase Inhibitors with Unusual Binding Modes
J.Med.Chem., 55, 2012
|
|
3BHY
 
 | | Crystal structure of human death associated protein kinase 3 (DAPK3) in complex with a beta-carboline ligand | | Descriptor: | (4R)-7,8-dichloro-1',9-dimethyl-1-oxo-1,2,4,9-tetrahydrospiro[beta-carboline-3,4'-piperidine]-4-carbonitrile, CHLORIDE ION, Death-associated protein kinase 3 | | Authors: | Filippakopoulos, P, Rellos, P, Eswaran, J, Fedorov, O, Berridge, G, Niesen, F, Bracher, F, Huber, K, Pike, A.C.W, Roos, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-29 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | 7,8-dichloro-1-oxo-beta-carbolines as a versatile scaffold for the development of potent and selective kinase inhibitors with unusual binding modes.
J.Med.Chem., 55, 2012
|
|
2WU7
 
 | | Crystal Structure of the Human CLK3 in complex with V25 | | Descriptor: | CHLORIDE ION, DUAL SPECIFICITY PROTEIN KINASE CLK3, SULFATE ION, ... | | Authors: | Muniz, J.R.C, Fedorov, O, King, O, Filippakopoulos, P, Bullock, A.N, Phillips, C, Heightman, T, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Bracher, F, Huber, K, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S. | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Specific Clk Inhibitors from a Novel Chemotype for Regulation of Alternative Splicing.
Chem.Biol, 18, 2011
|
|
8OU2
 
 | | Crystal structure of the bromodomain of human Bromodomain and PHD finger-containing Transcription Factor (BPTF) bound to a potent inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 7-(1-cyclopropylpyrazol-4-yl)-2-[[2-fluoranyl-4-(4-methylpiperazin-1-yl)sulfonyl-phenyl]amino]-3-methyl-pyrido[1,2-a]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Balikci, E, Ni, X, Bountra, C, von Delft, F, Huber, K. | | Deposit date: | 2023-04-21 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the bromodomain of human Bromodomain and PHD finger-containing Transcription Factor (BPTF) bound to a potent inhibitor
To Be Published
|
|
6S5H
 
 | | Structure of the human RAB38 in complex with GTP | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Diaz-Saez, L, Jung, S, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-07-01 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the human RAB38 in complex with GTP
To Be Published
|
|
6S5F
 
 | | Structure of the human RAB39B in complex with GMPPNP | | Descriptor: | 1,2-ETHANEDIOL, GLYCINE, MAGNESIUM ION, ... | | Authors: | Diaz-Saez, L, Jung, S, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-07-01 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the human RAB39B in complex with GMPPNP
To Be Published
|
|
5MQ1
 
 | | Crystal structure of the BRD7 bromodomain in complex with BI-9564 | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-[(dimethylamino)methyl]-2,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, Bromodomain-containing protein 7, ... | | Authors: | Diaz-Saez, L, Martin, L.J, Panagakou, I, Picaud, S, Krojer, T, von Delft, F, Knapp, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-12-19 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the BRD7 bromodomain in complex with BI-9564
To Be Published
|
|
5MP0
 
 | | Human m7GpppN-mRNA Hydrolase (DCP2, NUDT20) Catalytic Domain | | Descriptor: | 1,2-ETHANEDIOL, m7GpppN-mRNA hydrolase | | Authors: | Mathea, S, Salah, E, Velupillai, S, Tallant, C, Pike, A.C.W, Bushell, S.R, Faust, B, Wang, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Huber, K. | | Deposit date: | 2016-12-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Human m7GpppN-mRNA Hydrolase (DCP2, NUDT20) Catalytic Domain
To Be Published
|
|
8BI5
 
 | | Crystal structure of human Choline Kinase A in complex with UNC0737 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclohexyl-6-methoxy-~{N}-methyl-~{N}-(1-propan-2-ylpiperidin-4-yl)-7-(3-pyrrolidin-1-ylpropoxy)quinazolin-4-amine, CHLORIDE ION, ... | | Authors: | Diaz-Saez, L, Ward, J, Kennedy, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human Choline Kinase A in complex with UNC0737
To Be Published
|
|
8BI6
 
 | | Crystal structure of human Choline Kinase A in complex with UNC0638 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclohexyl-6-methoxy-N-[1-(1-methylethyl)piperidin-4-yl]-7-(3-pyrrolidin-1-ylpropoxy)quinazolin-4-amine, CHLORIDE ION, ... | | Authors: | Diaz-Saez, L, Ward, J, Kennedy, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human Choline Kinase A in complex with UNC0638
To Be Published
|
|
5LTU
 
 | | Crystal Structure of NUDT4A- Diphosphoinositol polyphosphate phosphohydrolase 2 | | Descriptor: | 1,2-ETHANEDIOL, Diphosphoinositol polyphosphate phosphohydrolase 2 | | Authors: | Srikannathasan, V, Nunez, C.A, Tallant, C, Siejka, P, Mathea, S, Newman, J, Strain-Damerell, C, Elkins, J.M, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Huber, K. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal Structure of Human NUDT4A- Diphosphoinositol polyphosphate phosphohydrolase 2
To Be Published
|
|
5LF9
 
 | | Crystal structure of human NUDT22 | | Descriptor: | Nucleoside diphosphate-linked moiety X motif 22 | | Authors: | Tallant, C, Siejka, P, Mathea, S, Shrestha, L, Krojer, T, Srikannathasan, V, Elkins, J.M, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-30 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of human NUDT22
To Be Published
|
|
6FDY
 
 | | Unc-51-Like Kinase 3 (ULK3) In Complex With Bosutinib | | Descriptor: | 4-[(2,4-dichloro-5-methoxyphenyl)amino]-6-methoxy-7-[3-(4-methylpiperazin-1-yl)propoxy]quinoline-3-carbonitrile, Serine/threonine-protein kinase ULK3 | | Authors: | Mathea, S, Salah, E, Moroglu, M, Scorah, A, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Knapp, S. | | Deposit date: | 2017-12-27 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unc-51-Like Kinase 3 (ULK3) In Complex With Bosutinib
To Be Published
|
|
6FDZ
 
 | | Unc-51-Like Kinase 3 (ULK3) In Complex With Momelotinib | | Descriptor: | Momelotinib, Serine/threonine-protein kinase ULK3 | | Authors: | Mathea, S, Salah, E, Moroglu, M, Scorah, A, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Knapp, S. | | Deposit date: | 2017-12-27 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Unc-51-Like Kinase 3 (ULK3) In Complex With Momelotinib
To Be Published
|
|
6GRU
 
 | | Crystal structure of human NUDT5 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Fairhead, M, MacLean, E, Diaz Saez, L, Strain-Damerell, C, Elkins, J, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-12 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of human NUDT5
To Be Published
|
|
5QGS
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with FMOPL000476a | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, N-(4-chlorophenyl)-N'-[(2R)-1-hydroxybutan-2-yl]urea, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2018-05-15 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QH6
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with NUOOA000259a | | Descriptor: | 2-(3-hydroxyphenyl)-N-(pyridin-2-yl)acetamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2018-05-15 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QGY
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with NUOOA000158 | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, N-(3-chlorophenyl)-2-phenylacetamide, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2018-05-15 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
