2K60
 
 | | NMR structure of calcium-loaded STIM1 EF-SAM | | Descriptor: | CALCIUM ION, PROTEIN (Stromal interaction molecule 1) | | Authors: | Stathopulos, P.B, Ikura, M. | | Deposit date: | 2008-07-02 | | Release date: | 2008-10-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and mechanistic insights into STIM1-mediated initiation of store-operated calcium entry.
Cell(Cambridge,Mass.), 135, 2008
|
|
2L5Y
 
 | |
3BS7
 
 | |
3BQ7
 
 | |
2B6G
 
 | |
2D3D
 
 | | crystal structure of the RNA binding SAM domain of saccharomyces cerevisiae Vts1 | | Descriptor: | CALCIUM ION, Vts1 protein | | Authors: | Aviv, T, Amborski, A.N, Zhao, X.S, Kwan, J.J, Johnson, P.E, Sicheri, F, Donaldson, L.W. | | Deposit date: | 2005-09-27 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The NMR and X-ray Structures of the Saccharomyces cerevisiae Vts1 SAM Domain Define a Surface for the Recognition of RNA Hairpins
J.Mol.Biol., 356, 2006
|
|
2DKY
 
 | | Solution structure of the SAM-domain of Rho-GTPase-activating protein 7 | | Descriptor: | Rho-GTPase-activating protein 7 | | Authors: | Goroncy, A.K, Sato, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-14 | | Release date: | 2007-04-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAM-domain of Rho-GTPase-activating protein 7
To be Published
|
|
2DL0
 
 | | Solution structure of the SAM-domain of the SAM and SH3 domain containing protein 1 | | Descriptor: | SAM and SH3 domain-containing protein 1 | | Authors: | Goroncy, A.K, Sato, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-14 | | Release date: | 2006-10-14 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAM-domain of the SAM and SH3 domain containing protein 1
To be Published
|
|
2E8O
 
 | | Solution structure of the N-terminal SAM-domain of the SAM domain and HD domain containing protein 1 (Dendritic cell-derived IFNG-induced protein) (DCIP) (Monocyte protein 5) (MOP-5) | | Descriptor: | SAM domain and HD domain-containing protein 1 | | Authors: | Goroncy, A.K, Tochio, N, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-22 | | Release date: | 2007-07-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal SAM-domain of the SAM domain and HD domain containing protein 1 (Dendritic cell-derived IFNG-induced protein) (DCIP) (Monocyte protein 5) (MOP-5)
To be Published
|
|
2EAP
 
 | | Solution structure of the N-terminal SAM-domain of human lymphocyte cytosolic protein 2 | | Descriptor: | Lymphocyte cytosolic protein 2 | | Authors: | Goroncy, A.K, Sato, M, Tochio, N, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-31 | | Release date: | 2008-02-05 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal SAM-domain of human lymphocyte cytosolic protein 2
To be Published
|
|
2ESE
 
 | | Structure of the SAM domain of Vts1p in complex with RNA | | Descriptor: | 5'-R(*GP*GP*AP*GP*AP*GP*GP*CP*UP*CP*UP*GP*GP*CP*AP*GP*CP*UP*UP*UP*UP*CP*C)-3', Vts1p | | Authors: | Allain, F.H.T. | | Deposit date: | 2005-10-26 | | Release date: | 2006-01-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Shape-specific recognition in the structure of the Vts1p SAM domain with RNA.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2ES6
 
 | | Structure of the SAM domain of Vts1p | | Descriptor: | Vts1p | | Authors: | Allain, F.H.T. | | Deposit date: | 2005-10-25 | | Release date: | 2006-01-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Shape-specific recognition in the structure of the Vts1p SAM domain with RNA.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2F8K
 
 | | Sequence specific recognition of RNA hairpins by the SAM domain of Vts1 | | Descriptor: | 5'-R(*UP*AP*AP*UP*CP*UP*UP*UP*GP*AP*CP*AP*GP*AP*UP*U)-3', Protein VTS1 | | Authors: | Aviv, T, Lin, Z, Ben-Ari, G, Smibert, C.A, Sicheri, F. | | Deposit date: | 2005-12-02 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sequence-specific recognition of RNA hairpins by the SAM domain of Vts1p.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2FE9
 
 | |
2GLE
 
 | |
2GYT
 
 | |
2H80
 
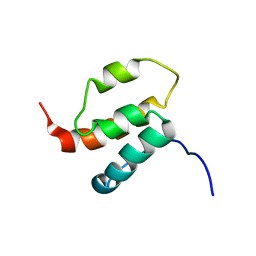 | | NMR structures of SAM domain of Deleted in Liver Cancer 2 (DLC2) | | Descriptor: | StAR-related lipid transfer protein 13 | | Authors: | Li, H.Y, Fung, K.L, Jin, D.Y, Chung, S.S, Ko, B.C, Sun, H.Z. | | Deposit date: | 2006-06-06 | | Release date: | 2007-05-15 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structures, dynamics, and lipid-binding of the sterile alpha-motif domain of the deleted in liver cancer 2
Proteins, 67, 2007
|
|
4TWN
 
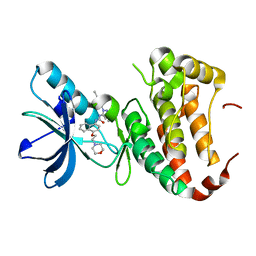 | | Human EphA3 Kinase domain in complex with Birb796 | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Ephrin type-A receptor 3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-07-01 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.706 Å) | | Cite: | Structural Analysis of the Binding of Type I, I1/2, and II Inhibitors to Eph Tyrosine Kinases.
Acs Med.Chem.Lett., 6, 2015
|
|
7DJT
 
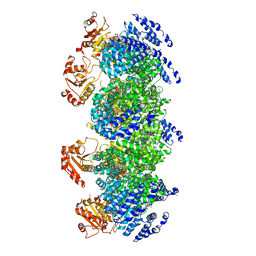 | | Human SARM1 inhibitory state bounded with inhibitor dHNN | | Descriptor: | NAD(+) hydrolase SARM1, O3-methyl O5-(2-methylpropyl) 2,6-dimethyl-4-[2-(oxidanylamino)phenyl]pyridine-3,5-dicarboxylate | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2020-11-21 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Permeant fluorescent probes visualize the activation of SARM1 and uncover an anti-neurodegenerative drug candidate.
Elife, 10, 2021
|
|
7QG0
 
 | | Inhibitor-induced hSARM1 duplex | | Descriptor: | NAD(+) hydrolase SARM1 | | Authors: | Zalk, R, Kahzma, T, Guez-Haddad, J. | | Deposit date: | 2021-12-07 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | A duplex structure of SARM1 octamers stabilized by a new inhibitor.
Cell.Mol.Life Sci., 80, 2022
|
|
8GQ5
 
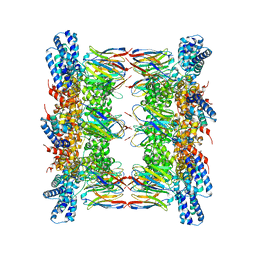 | |
8GNJ
 
 | | Human SARM1 bounded with NMN and Nanobody-C6, Conformation 2 | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1, Nanobody-C6 | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | A conformation-specific nanobody targeting the nicotinamide mononucleotide-activated state of SARM1.
Nat Commun, 13, 2022
|
|
8GNI
 
 | | Human SARM1 bounded with NMN and Nanobody-C6, Conformation 1 | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1, Nanobody C6 | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | A conformation-specific nanobody targeting the nicotinamide mononucleotide-activated state of SARM1.
Nat Commun, 13, 2022
|
|
8P2L
 
 | |
7LD0
 
 | | Cryo-EM structure of ligand-free Human SARM1 | | Descriptor: | NAD(+) hydrolase SARM1 | | Authors: | Nanson, J.D, Gu, W, Luo, Z, Jia, X, Landsberg, M.J, Kobe, B, Ve, T. | | Deposit date: | 2021-01-12 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
