5VFB
 
 | | 1.36 Angstrom Resolution Crystal Structure of Malate Synthase G from Pseudomonas aeruginosa in Complex with Glycolic Acid. | | Descriptor: | CHLORIDE ION, GLYCOLIC ACID, Malate synthase G, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-07 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | 1.36 Angstrom Resolution Crystal Structure of Malate Synthase G from Pseudomonas aeruginosa in Complex with Glycolic Acid.
To Be Published
|
|
5YVT
 
 | |
3MHU
 
 | | Crystal structure of dihydroorotate dehydrogenase from Leishmania major in complex with 5-Nitroorotic acid | | Descriptor: | 5-nitro-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Pinheiro, M.P, Rocha, J.R, Cheleski, J, Montanari, C.A, Nonato, M.C. | | Deposit date: | 2010-04-08 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Novel insights for dihydroorotate dehydrogenase class 1A inhibitors discovery.
Eur.J.Med.Chem., 45, 2010
|
|
8II0
 
 | | FACTOR INHIBITING HIF-1 ALPHA in complex with (5-(3-(3-chlorophenyl)isoxazol-5-yl)-3-hydroxypicolinoyl)glycine | | Descriptor: | 2-[[5-[3-(3-chlorophenyl)-1,2-oxazol-5-yl]-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Corner, T, Zhang, X, Schofield, C.J. | | Deposit date: | 2023-02-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | A Small-Molecule Inhibitor of Factor Inhibiting HIF Binding to a Tyrosine-Flip Pocket for the Treatment of Obesity.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
1M56
 
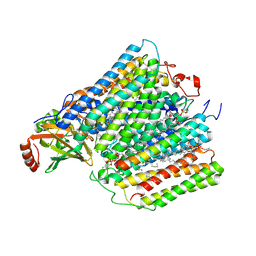 | | Structure of cytochrome c oxidase from Rhodobactor sphaeroides (Wild Type) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Svensson-Ek, M, Abramson, J, Larsson, G, Tornroth, S, Brezezinski, P, Iwata, S. | | Deposit date: | 2002-07-08 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides.
J.Mol.Biol., 321, 2002
|
|
4KWG
 
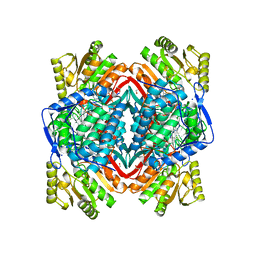 | | Crystal Structure Analysis of ALDH2+ALDiB13 | | Descriptor: | 1,2-ETHANEDIOL, 7-bromo-5-methyl-1H-indole-2,3-dione, Aldehyde dehydrogenase, ... | | Authors: | Hurley, T.D, Kimble-Hill, A.C. | | Deposit date: | 2013-05-24 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development of selective inhibitors for aldehyde dehydrogenases based on substituted indole-2,3-diones.
J.Med.Chem., 57, 2014
|
|
5JK4
 
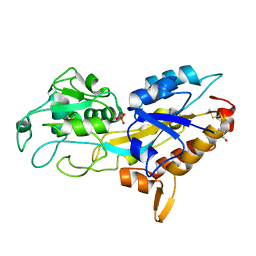 | | Phosphate-Binding Protein from Stenotrophomonas maltophilia. | | Descriptor: | Alkaline phosphatase, PHOSPHATE ION | | Authors: | Keegan, R, Waterman, D, Hopper, D, Coates, L, Guo, J, Coker, A.R, Erskine, P.T, Wood, S.P, Cooper, J.B. | | Deposit date: | 2016-04-25 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1 angstrom resolution structure of a periplasmic phosphate-binding protein from Stenotrophomonas maltophilia: a crystallization contaminant identified by molecular replacement using the entire Protein Data Bank.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1M57
 
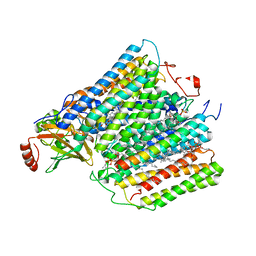 | | Structure of cytochrome c oxidase from Rhodobacter sphaeroides (EQ(I-286) mutant)) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Svensson-Ek, M, Abramson, J, Larsson, G, Tornroth, S, Brezezinski, P, Iwata, S. | | Deposit date: | 2002-07-08 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides.
J.Mol.Biol., 321, 2002
|
|
6LVR
 
 | |
5CID
 
 | |
6TDU
 
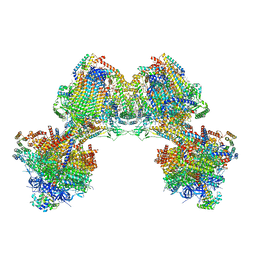 | | Cryo-EM structure of Euglena gracilis mitochondrial ATP synthase, full dimer, rotational states 1 | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Muhleip, A, Amunts, A. | | Deposit date: | 2019-11-10 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Structure of a mitochondrial ATP synthase with bound native cardiolipin.
Elife, 8, 2019
|
|
8I5B
 
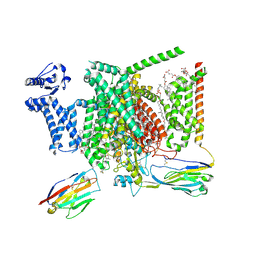 | | Structure of human Nav1.7 in complex with bupivacaine | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-01-24 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
3K8T
 
 | | Structure of eukaryotic rnr large subunit R1 complexed with designed adp analog compound | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 2'-deoxy-2'-(2-hydroxyethyl)adenosine 5'-(trihydrogen diphosphate), GLYCEROL, ... | | Authors: | Sun, D, Xu, H, Dealwis, C, Lee, R.E. | | Deposit date: | 2009-10-14 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design, Synthesis, and Evaluation of 2'-(2-Hydroxyethyl)-2'-deoxyadenosine and the 5'-Diphosphate Derivative as Ribonucleotide Reductase Inhibitors
Chemmedchem, 4, 2009
|
|
6SPA
 
 | | A4V MUTANT OF HUMAN SUPEROXIDE DISMUTASE 1 IN C2 SPACE GROUP | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Shahid, M, Chantadul, V, Amporndanai, K, Wright, G, Antonyuk, S, Hasnain, S. | | Deposit date: | 2019-08-31 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Ebselen as template for stabilization of A4V mutant dimer for motor neuron disease therapy.
Commun Biol, 3, 2020
|
|
5ODC
 
 | | Heterodisulfide reductase / [NiFe]-hydrogenase complex from Methanothermococcus thermolithotrophicus at 2.3 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETATE ION, ... | | Authors: | Wagner, T, Koch, J, Ermler, U, Shima, S. | | Deposit date: | 2017-07-05 | | Release date: | 2017-08-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction.
Science, 357, 2017
|
|
6AOH
 
 | | Crystal structure of Toxoplasma gondii TS-DHFR complexed with NADPH, dUMP, PDDF, and 5-(4-(3-(2-methoxypyrimidin-5-yl)phenyl)piperazin-1-yl)pyrimidine-2,4-diamine (TRC-2533) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-{4-[3-(2-methoxypyrimidin-5-yl)phenyl]piperazin-1-yl}pyrimidine-2,4-diamine, ... | | Authors: | Thomas, S.B, Li, Y, Chen, Z, Lu, H. | | Deposit date: | 2017-08-16 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Identification of selective, brain penetrant Toxoplasma gondii dihydrofolate reductase inhibitors for the treatment of toxoplasmosis
To Be Published
|
|
5I9X
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with bosutinib (SKI-606) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2,4-dichloro-5-methoxyphenyl)amino]-6-methoxy-7-[3-(4-methylpiperazin-1-yl)propoxy]quinoline-3-carbonitrile, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.427 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
5CG2
 
 | | Crystal structure of E. coli FabI bound to the thiocarbamoylated benzodiazaborine inhibitor 35b. | | Descriptor: | 1-hydroxy-2,3,1-benzodiazaborinine-2(1H)-carbothioamide, Enoyl-[acyl-carrier-protein] reductase [NADH] FabI, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jordan, C.A, Vey, J.L. | | Deposit date: | 2015-07-09 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystallographic insights into the structure-activity relationships of diazaborine enoyl-ACP reductase inhibitors.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5U29
 
 | | Crystal structure of Cryptococcus neoformans H99 Acetyl-CoA Synthetase in complex with Ac-AMS | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-(acetylsulfamoyl)adenosine, Acetyl-coenzyme A synthetase, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Fox III, D, Edwards, T.E, Potts, K.T, Taylor, B.M. | | Deposit date: | 2016-11-30 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Cryptococcus neoformans H99 Acetyl-CoA Synthetase in complex with Ac-AMS
To Be Published
|
|
6FXJ
 
 | | Structure of coproheme decarboxylase from Listeria monocytogenes in complex with iron coproporphyrin III | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, CHLORIDE ION, N-PROPANOL, ... | | Authors: | Hofbauer, S, Pfanzagl, V, Mlynek, G. | | Deposit date: | 2018-03-09 | | Release date: | 2019-07-10 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Redox Cofactor Rotates during Its Stepwise Decarboxylation: Molecular Mechanism of Conversion of Coproheme to Hemeb.
Acs Catalysis, 9, 2019
|
|
6MTU
 
 | | Crystal structure of human Scribble PDZ1:pMCC complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Colorectal mutant cancer protein, ... | | Authors: | Caria, S, Stewart, B.Z, Humbert, P.O, Kvansakul, M. | | Deposit date: | 2018-10-22 | | Release date: | 2019-08-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structural analysis of phosphorylation-associated interactions of human MCC with Scribble PDZ domains.
Febs J., 286, 2019
|
|
6MUQ
 
 | | 1.67 Angstrom Resolution Crystal Structure of Murein-DD-endopeptidase from Yersinia enterocolitica. | | Descriptor: | ACETATE ION, Murein-DD-endopeptidase, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-23 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | 1.67 Angstrom Resolution Crystal Structure of Murein-DD-endopeptidase from Yersinia enterocolitica.
To Be Published
|
|
7Y00
 
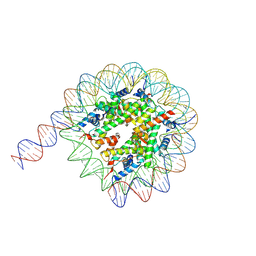 | | Cryo-EM structure of the nucleosome containing 169 base-pair DNA with a p53 target sequence | | Descriptor: | DNA (169-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
7XZY
 
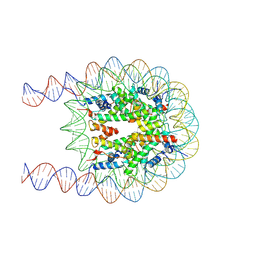 | | Cryo-EM structure of the nucleosome containing 193 base-pair DNA with a p53 target sequence | | Descriptor: | DNA (193-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
7XZZ
 
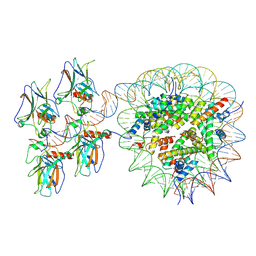 | | Cryo-EM structure of the nucleosome in complex with p53 | | Descriptor: | Cellular tumor antigen p53, DNA (169-MER), Histone H2A type 1-B/E, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
