3RLH
 
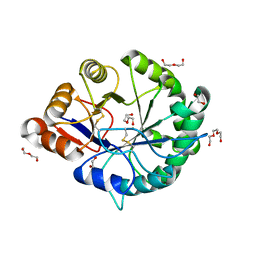 | | Crystal structure of a class II phospholipase D from Loxosceles intermedia venom | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Giuseppe, P.O, Ullah, A, Veiga, S.S, Murakami, M.T, Arni, R.K. | | Deposit date: | 2011-04-19 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure of a novel class II phospholipase D: Catalytic cleft is modified by a disulphide bridge.
Biochem.Biophys.Res.Commun., 409, 2011
|
|
3RM5
 
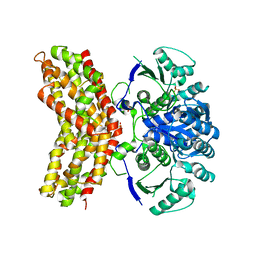 | |
3R2Y
 
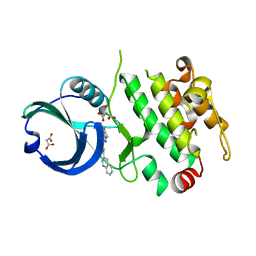 | | MK2 kinase bound to Compound 1 | | Descriptor: | 2-(2-QUINOLIN-3-YLPYRIDIN-4-YL)-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MALONATE ION, MAP kinase-activated protein kinase 2 | | Authors: | Oubrie, A, Leonard, P. | | Deposit date: | 2011-03-15 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based lead identification of ATP-competitive MK2 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3RMT
 
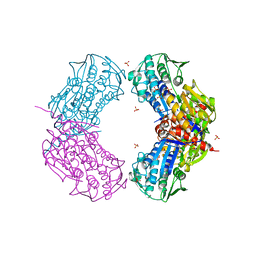 | | Crystal structure of putative 5-enolpyruvoylshikimate-3-phosphate synthase from Bacillus halodurans C-125 | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase 1, SULFATE ION | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Ramagopal, U, Zencheck, W, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-04-21 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of putative 5-enolpyruvoylshikimate-3-phosphate synthase from Bacillus halodurans C-125
To be Published
|
|
3RO7
 
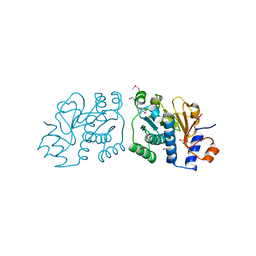 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with Thymine. | | Descriptor: | Apolipoprotein A-I-binding protein, SULFATE ION, THYMINE | | Authors: | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2011-04-25 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3R3U
 
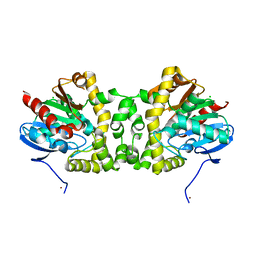 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - WT/apo | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, NICKEL (II) ION | | Authors: | Chan, P.W.Y, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-03-16 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mapping the reaction coordinates of enzymatic defluorination.
J.Am.Chem.Soc., 133, 2011
|
|
3RPN
 
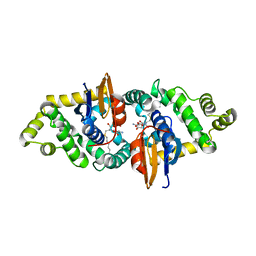 | | Crystal structure of human kappa class glutathione transferase in complex with S-hexylglutathione | | Descriptor: | Glutathione S-transferase kappa 1, S-HEXYLGLUTATHIONE | | Authors: | Wang, B, Peng, Y, Zhang, T, Ding, J. | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures and kinetic studies of human Kappa class glutathione transferase provide insights into the catalytic mechanism.
Biochem.J., 439, 2011
|
|
3R6R
 
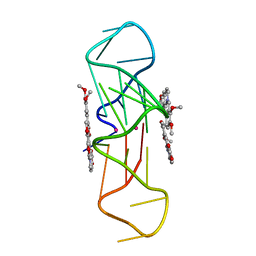 | | Structure of the complex of an intramolecular human telomeric DNA with Berberine formed in K+ solution | | Descriptor: | BERBERINE, DNA (22-mer), POTASSIUM ION | | Authors: | Ferraroni, M, Bazzicalupi, C, Gratteri, P, Bilia, A.R. | | Deposit date: | 2011-03-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of human telomeric DNA complexed with berberine: an interesting case of stacked ligand to G-tetrad ratio higher than 1:1.
Nucleic Acids Res., 41, 2013
|
|
3RSF
 
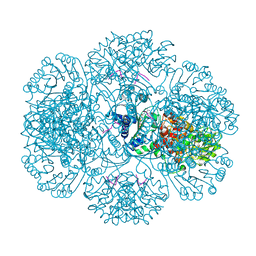 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with P1,P4-Di(adenosine-5') tetraphosphate | | Descriptor: | BIS(ADENOSINE)-5'-TETRAPHOSPHATE, POTASSIUM ION, Putative uncharacterized protein, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3PZ5
 
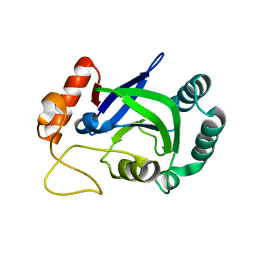 | | The crystal structure of AaLeuRS-CP1-D20 | | Descriptor: | Leucyl-tRNA synthetase subunit alpha | | Authors: | Liu, R.J, Wang, E.D. | | Deposit date: | 2010-12-14 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Peripheral insertion modulates the editing activity of the isolated CP1 domain of leucyl-tRNA synthetase
Biochem.J., 440, 2011
|
|
3PZP
 
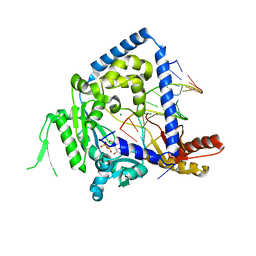 | | Human DNA polymerase kappa extending opposite a cis-syn thymine dimer | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*CP*A)-3', 5'-D(*TP*TP*CP*CP*(TTD)P*GP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Vasquez-Del Carpio, R, Silverstein, T.D, Lone, S, Johnson, R.E, Prakash, S, Prakash, L, Aggarwal, A.K. | | Deposit date: | 2010-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.336 Å) | | Cite: | Role of human DNA polymerase kappa in extension opposite from a cis-syn thymine dimer.
J.Mol.Biol., 408, 2011
|
|
3Q0J
 
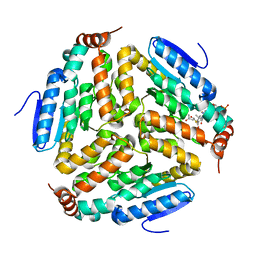 | |
3Q17
 
 | | Structure of a slow CLC Cl-/H+ antiporter from a cyanobacterium in Bromide | | Descriptor: | BROMIDE ION, Sll0855 protein | | Authors: | Jayaram, H, Robertson, J.L, Fang, W, Williams, C, Miller, C. | | Deposit date: | 2010-12-16 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of a Slow CLC Cl(-)/H(+) Antiporter from a Cyanobacterium.
Biochemistry, 50, 2011
|
|
3PKY
 
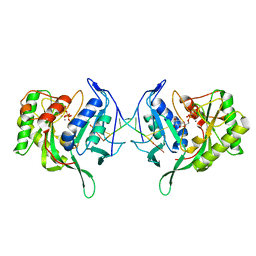 | | Polymerase Domain from Mycobacterium tuberculosis Ligase D in complex with DNA, UTP and Manganese. | | Descriptor: | DNA 5'-D(*G*CP*CP*GP*CP*AP*AP*CP*GP*CP*AP*CP*G)-3', DNA 5'-D(P*GP*CP*GP*GP*C)-3', MANGANESE (II) ION, ... | | Authors: | Brissett, N.C, Fox, G.C, Pitcher, R.S, Doherty, A.J. | | Deposit date: | 2010-11-12 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of a Preternary Complex Involving a Prokaryotic NHEJ DNA Polymerase.
Mol.Cell, 41, 2011
|
|
3Q29
 
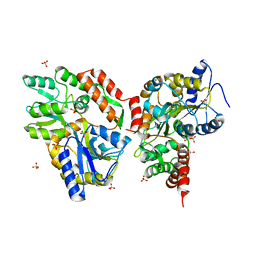 | | Cyrstal structure of human alpha-synuclein (1-19) fused to maltose binding protein (MBP) | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein/alpha-synuclein chimeric protein, SULFATE ION, ... | | Authors: | Zhao, M, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2010-12-19 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of segments of alpha-synuclein fused to maltose-binding protein suggest intermediate states during amyloid formation
Protein Sci., 20, 2011
|
|
3Q4L
 
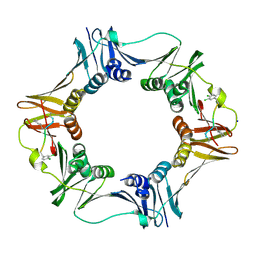 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, SODIUM ION, peptide ligand | | Authors: | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-12-28 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
3PJY
 
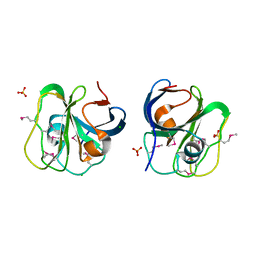 | |
3PNQ
 
 | | Crystal Structure of E.coli Dha kinase DhaK (H56N) complex with Dha | | Descriptor: | Dihydroxyacetone, PTS-dependent dihydroxyacetone kinase, dihydroxyacetone-binding subunit dhaK | | Authors: | Shi, R, McDonald, L, Matte, A, Cygler, M, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-11-19 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PM6
 
 | |
3PN5
 
 | |
3PR3
 
 | | Crystal structure of Plasmodium falciparum glucose-6-phosphate isomerase (PF14_0341) in complex with fructose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Glucose-6-phosphate isomerase, PHOSPHATE ION | | Authors: | Gileadi, T, Wernimont, A.K, Hutchinson, A, Weadge, J, Cossar, D, Lew, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Hills, T, Pizarro, J.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-29 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Plasmodium falciparum glucose-6-phosphate isomerase (PF14_0341) in complex with fructose-6-phosphate
To be Published
|
|
3PO1
 
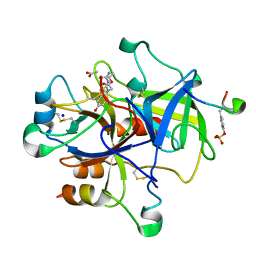 | | Thrombin in complex with Benzothiazole Guanidine | | Descriptor: | ACETATE ION, SODIUM ION, Thrombin heavy chain, ... | | Authors: | Xue, Y. | | Deposit date: | 2010-11-21 | | Release date: | 2011-11-23 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of benzothiazole guanidines as novel inhibitors of thrombin and trypsin IV.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3PSK
 
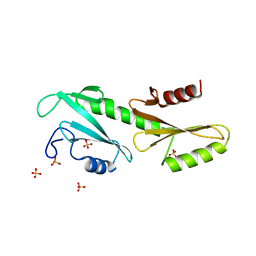 | |
3PPW
 
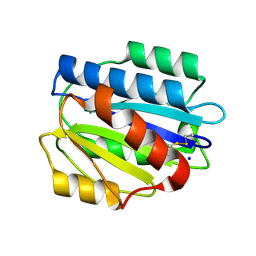 | | Crystal structure of the D1596A mutant of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | SODIUM ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
3PQ7
 
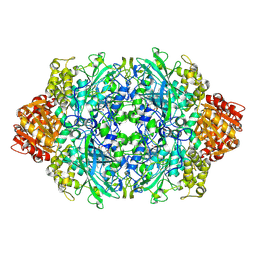 | | Structure of I274C variant of E. coli KatE[] - Images 31-36 | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
