5UCY
 
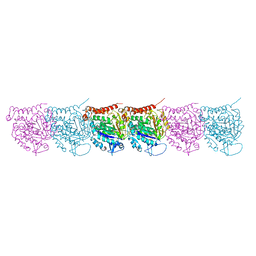 | | Cryo-EM map of protofilament of microtubule doublet | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ichikawa, M, Liu, D, Kastritis, P.L, Basu, K, Bui, K.H. | | Deposit date: | 2016-12-22 | | Release date: | 2017-05-10 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Subnanometre-resolution structure of the doublet microtubule reveals new classes of microtubule-associated proteins.
Nat Commun, 8, 2017
|
|
8U06
 
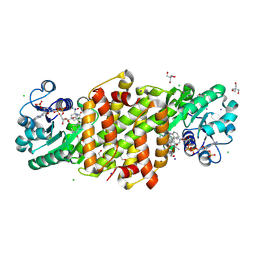 | |
7UPS
 
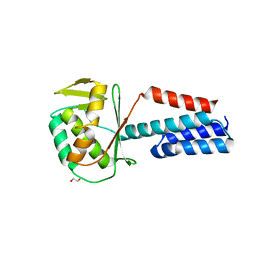 | |
8U07
 
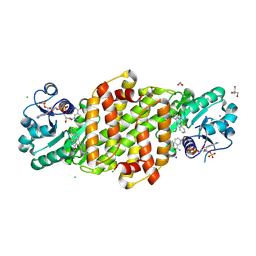 | |
7UBX
 
 | |
7UBY
 
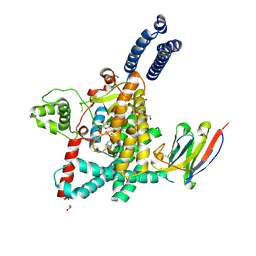 | | Structure of the GTD domain of Clostridium difficile toxin A in complex with VHH AH3 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Glucosyltransferase TcdA, ... | | Authors: | Chen, B, Rongsheng, J, Kay, P. | | Deposit date: | 2022-03-15 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Neutralizing epitopes on Clostridioides difficile toxin A revealed by the structures of two camelid VHH antibodies.
Front Immunol, 13, 2022
|
|
8U0H
 
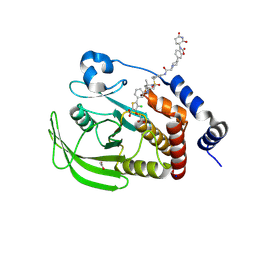 | | Crystal structure of PTPN2 with a PROTAC | | Descriptor: | (5P)-3-(carboxymethoxy)-4-chloro-5-(3-{[(4S)-1-({3-[2-(4-{3-[(3R)-2,6-dioxopiperidin-3-yl]-2-oxo-2,3-dihydro-1,3-benzoxazol-6-yl}piperidin-1-yl)acetamido]phenyl}methanesulfonyl)-2,2-dimethylpiperidin-4-yl]amino}phenyl)thiophene-2-carboxylic acid, ACETATE ION, PTPN2, ... | | Authors: | Jain, R, Longenecker, K, Qiu, W. | | Deposit date: | 2023-08-29 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mechanistic insights into a heterobifunctional degrader-induced PTPN2/N1 complex.
Commun Chem, 7, 2024
|
|
7XMA
 
 | | Crystal structure of Bovine heart cytochrome c oxidase, apo structure with DMSO | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Nishida, Y, Shinzawa-Itoh, K, Mizuno, N, Kumasaka, T, Yoshikawa, S, Tsukihara, T, Takashima, S, Shintani, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
7WZX
 
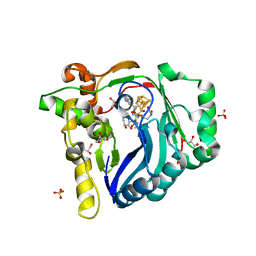 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | Descriptor: | (2~{S},4~{S},6~{R})-2-[(2~{S},3~{R},5~{S},6~{R})-3,5-bis(methylamino)-2,4,6-tris(oxidanyl)cyclohexyl]oxy-6-methyl-4-oxidanyl-oxan-3-one, 4Fe-4S cluster-binding domain-containing protein, GLYCEROL, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2022-02-19 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.980013 Å) | | Cite: | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7XMB
 
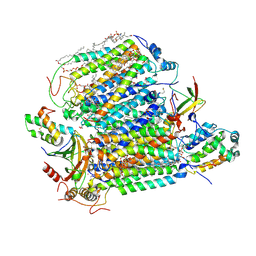 | | Crystal structure of Bovine heart cytochrome c oxidase, the structure complexed with an allosteric inhibitor T113 | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Nishida, Y, Shinzawa-Itoh, K, Mizuno, N, Kumasaka, T, Yoshikawa, S, Tsukihara, T, Shintani, Y, Takashima, S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
7WZV
 
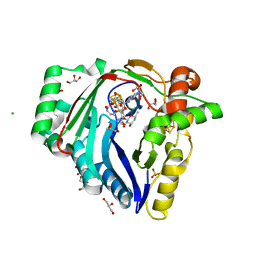 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | Descriptor: | (1~{S},2~{R},4~{S},5~{R})-2,4-bis(methylamino)-6-[(2~{S},3~{R},4~{S},6~{R})-6-methyl-3,4-bis(oxidanyl)oxan-2-yl]oxy-cyclohexane-1,3,5-triol, 1,2-ETHANEDIOL, 4Fe-4S cluster-binding domain-containing protein, ... | | Authors: | Zhou, J.H, Hou, X.L. | | Deposit date: | 2022-02-19 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.899313 Å) | | Cite: | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7X0B
 
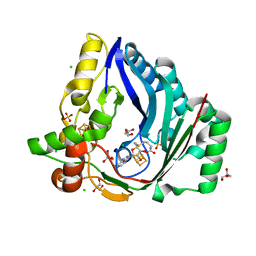 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | Descriptor: | 4Fe-4S cluster-binding domain-containing protein, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2022-02-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02027535 Å) | | Cite: | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
8DPY
 
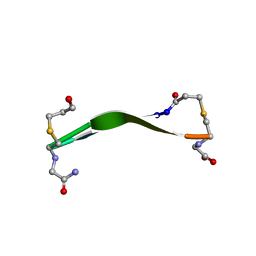 | |
7UFJ
 
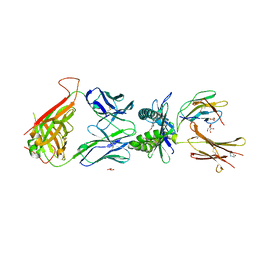 | | Structure of human MR1-ethylvanillin in complex with human MAIT A-F7 TCR | | Descriptor: | 3-ethoxy-4-hydroxybenzaldehyde, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Wang, C.J, Rossjohn, J, Le Nours, J. | | Deposit date: | 2022-03-22 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quantitative affinity measurement of small molecule ligand binding to Major Histocompatibility Complex class-I related protein 1 MR1.
J.Biol.Chem., 298, 2022
|
|
7UYG
 
 | |
7UYH
 
 | |
5TGA
 
 | |
7UXA
 
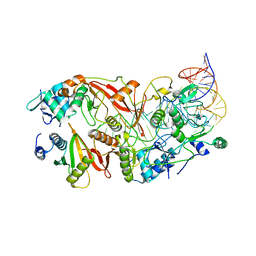 | | Human tRNA Splicing Endonuclease Complex bound to pre-tRNA-ARG | | Descriptor: | MAGNESIUM ION, RNA (78-MER), tRNA-splicing endonuclease subunit Sen15, ... | | Authors: | Stanley, R.E, Hayne, C.K. | | Deposit date: | 2022-05-05 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis for pre-tRNA recognition and processing by the human tRNA splicing endonuclease complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7N12
 
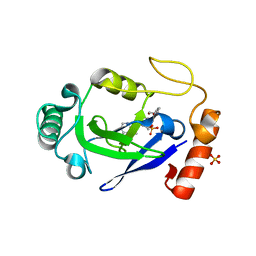 | | Crystal structure of the M. abscessus LeuRS editing domain in complex with epetraborole-AMP adduct | | Descriptor: | Leucine--tRNA ligase, SULFATE ION, [(1S,5R,6R,7'S,8R)-7'-(aminomethyl)-6-(6-aminopurin-9-yl)-2'-(3-oxidanylpropoxy)spiro[2,4,7-trioxa-3-boranuidabicyclo[3.3.0]octane-3,9'-8-oxa-9-boranuidabicyclo[4.3.0]nona-1(6),2,4-triene]-8-yl]methyl dihydrogen phosphate | | Authors: | Kalthoff, E, Schmeing, M. | | Deposit date: | 2021-05-26 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Efficacy of epetraborole against Mycobacterium abscessus is increased with norvaline.
Plos Pathog., 17, 2021
|
|
7N11
 
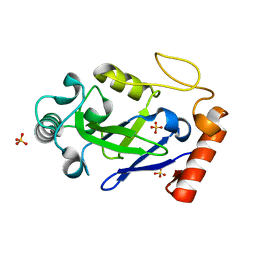 | |
5TGM
 
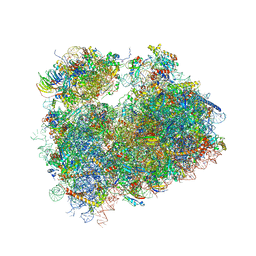 | | Crystal structure of the S.cerevisiae 80S ribosome in complex with the A-site bound aminoacyl-tRNA analog ACCA-Pro | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 3'-amino-3'-deoxyadenosine 5'-(dihydrogen phosphate), ... | | Authors: | Melnikov, S, Mailliot, J, Yusupov, M. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-18 | | Last modified: | 2022-12-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular insights into protein synthesis with proline residues.
EMBO Rep., 17, 2016
|
|
7N3K
 
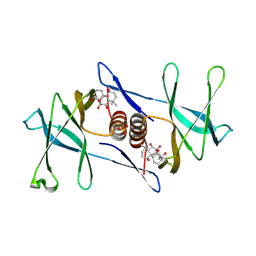 | | Oridonin-bound SARS-CoV-2 Nsp9 | | Descriptor: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, Non-structural protein 9, SULFATE ION | | Authors: | Littler, D.R, Gully, B.S, Rossjohn, J. | | Deposit date: | 2021-06-01 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A natural product compound inhibits coronaviral replication in vitro by binding to the conserved Nsp9 SARS-CoV-2 protein.
J.Biol.Chem., 297, 2021
|
|
8VVB
 
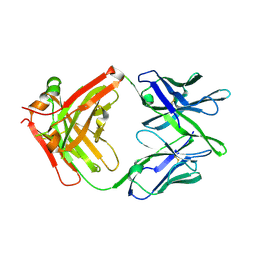 | | Influenza antibody L5A7 Fab | | Descriptor: | L5A7 Heavy Chain, L5A7 Light Chain | | Authors: | Harris, D.R, Olia, A.S, Kwong, P.D. | | Deposit date: | 2024-01-30 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Anti-idiotype isolation of a broad and potent influenza A virus-neutralizing human antibody.
Front Immunol, 15, 2024
|
|
7UR9
 
 | | SARS-Cov2 Main protease in complex with inhibitor CDD-1845 | | Descriptor: | (2P)-2-(isoquinolin-4-yl)-1-[4-(methylamino)-4-oxobutyl]-N-[(1S)-1-(naphthalen-2-yl)ethyl]-1H-benzimidazole-7-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lu, S, Palzkill, T. | | Deposit date: | 2022-04-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | DNA-encoded chemical libraries yield non-covalent and non-peptidic SARS-CoV-2 main protease inhibitors.
Commun Chem, 6, 2023
|
|
7URB
 
 | | Sars-Cov2 Main Protease in complex with CDD-1733 | | Descriptor: | (2P)-2-(isoquinolin-4-yl)-1-[(1s,3R)-3-(methylcarbamoyl)cyclobutyl]-N-{(1S)-1-[4-(trifluoromethyl)phenyl]butyl}-1H-benzimidazole-7-carboxamide, 3C-like proteinase | | Authors: | Lu, S, Palzkill, T, Matzuk, M.M, Judge, A. | | Deposit date: | 2022-04-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | DNA-encoded chemical libraries yield non-covalent and non-peptidic SARS-CoV-2 main protease inhibitors.
Commun Chem, 6, 2023
|
|
