3G8E
 
 | | Crystal Structure of Rattus norvegicus Visfatin/PBEF/Nampt in Complex with an FK866-based inhibitor | | Descriptor: | 3-[(1E)-3-oxo-3-({4-[1-(phenylcarbonyl)piperidin-4-yl]butyl}amino)prop-1-en-1-yl]-1-beta-D-ribofuranosylpyridinium, Nicotinamide phosphoribosyltransferase | | Authors: | Kang, G.B, Bae, M.H, Kim, M.K, Im, I, Kim, Y.C, Eom, S.H. | | Deposit date: | 2009-02-12 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Rattus norvegicus Visfatin/PBEF/Nampt in complex with an FK866-based inhibitor
Mol.Cells, 27, 2009
|
|
6NT3
 
 | | Cryo-EM structure of a human-cockroach hybrid Nav channel. | | Descriptor: | (7E,21R,24S)-27-amino-24-hydroxy-18,24-dioxo-19,23,25-trioxa-24lambda~5~-phosphaheptacos-7-en-21-yl (9Z,12E)-octadeca-9,12-dienoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Clairfeuille, T, Rohou, A, Payandeh, J. | | Deposit date: | 2019-01-28 | | Release date: | 2019-02-20 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of a human-cockroach hybrid Nav channel in the presence and absence of the alpha-scorpion toxin AaH2.
Science, 2019
|
|
5YV9
 
 | | Structure of CaMKK2 in complex with CKI-009 | | Descriptor: | 5-chloro-2-methoxy-4[(1Z)-3-(4-methoxyphenyl)-3-oxoprop-1-en-1-yl]aminobenzoic acid, CHLORIDE ION, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Niwa, H, Handa, N, Yokoyama, S. | | Deposit date: | 2017-11-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Protein ligand interaction analysis against new CaMKK2 inhibitors by use of X-ray crystallography and the fragment molecular orbital (FMO) method.
J.Mol.Graph.Model., 99, 2020
|
|
6O04
 
 | | M.tb MenD IntII bound with Inhibitor | | Descriptor: | (1~{R},2~{S},5~{S},6~{S})-2-[(1~{S})-1-[3-[(4-azanylidene-2-methyl-1~{H}-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl (phosphonooxy)phosphoryl]oxyethyl]-1,3-thiazol-3-ium-2-yl]-1,4-bis(oxidanyl)-4-oxidanylidene-butyl]-6-oxidanyl-5-(3-oxid anyl-3-oxidanylidene-prop-1-en-2-yl)oxy-cyclohex-3-ene-1-carboxylic acid, 1,4-dihydroxy-2-naphthoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, ... | | Authors: | Johnston, J.M, Bashiri, G, Bulloch, E.M, Jirgis, E.M.N, Nigon, L.V, Chuang, H, Baker, E.N. | | Deposit date: | 2019-02-15 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric regulation of menaquinone (vitamin K2) biosynthesis in the human pathogenMycobacterium tuberculosis.
J.Biol.Chem., 295, 2020
|
|
8DL3
 
 | | Crystal structure of the human queuine salvage enzyme DUF2419, complexed with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, Queuosine salvage protein | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-07-06 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
6NKM
 
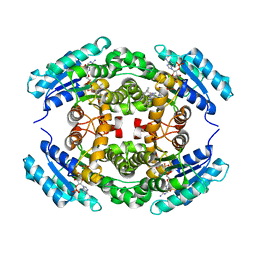 | | Structure of PhqE D166N Reductase/Diels-Alderase from Penicillium fellutanum in complex with NADP+ and substrate | | Descriptor: | 3-{[2-(2-methylbut-3-en-2-yl)-1H-indol-3-yl]methyl}-8H-pyrrolo[1,2-a]pyrazin-5-ium-1-olate, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short chain dehydrogenase | | Authors: | Newmister, S.A, Dan, Q, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-01-07 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Fungal indole alkaloid biogenesis through evolution of a bifunctional reductase/Diels-Alderase.
Nat.Chem., 11, 2019
|
|
3HKY
 
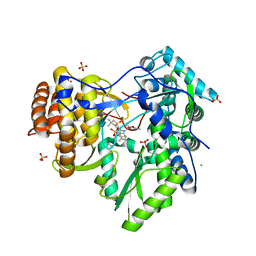 | | HCV NS5B polymerase genotype 1b in complex with 1,5 benzodiazepine 6 | | Descriptor: | (11S)-10-[(2,5-dimethyl-1,3-oxazol-4-yl)carbonyl]-11-{2-fluoro-4-[(2-methylprop-2-en-1-yl)oxy]phenyl}-3,3-dimethyl-2,3,4,5,10,11-hexahydrothiopyrano[3,2-b][1,5]benzodiazepin-6-ol 1,1-dioxide, CHLORIDE ION, RNA-directed RNA polymerase, ... | | Authors: | Nyanguile, O, De Bondt, H.L. | | Deposit date: | 2009-05-26 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1a/1b subtype profiling of nonnucleoside polymerase inhibitors of hepatitis C virus
J.Virol., 84, 2010
|
|
3HKW
 
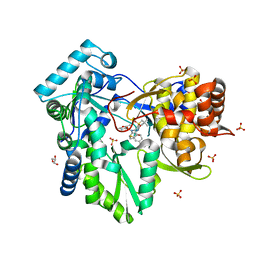 | | HCV NS5B genotype 1a in complex with 1,5 benzodiazepine inhibitor 6 | | Descriptor: | (11S)-10-[(2,5-dimethyl-1,3-oxazol-4-yl)carbonyl]-11-{2-fluoro-4-[(2-methylprop-2-en-1-yl)oxy]phenyl}-3,3-dimethyl-2,3,4,5,10,11-hexahydrothiopyrano[3,2-b][1,5]benzodiazepin-6-ol 1,1-dioxide, GLYCEROL, NS5B RNA-dependent RNA polymerase, ... | | Authors: | Nyanguile, O, De Bondt, H.L. | | Deposit date: | 2009-05-26 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1a/1b subtype profiling of nonnucleoside polymerase inhibitors of hepatitis C virus
J.Virol., 84, 2010
|
|
5YU9
 
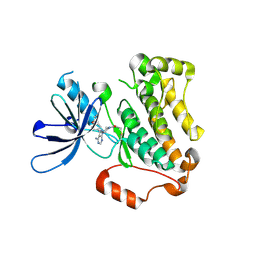 | | Crystal structure of EGFR 696-1022 T790M in complex with Ibrutinib | | Descriptor: | 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Yan, X.E, Yun, C.H. | | Deposit date: | 2017-11-21 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ibrutinib targets mutant-EGFR kinase with a distinct binding conformation.
Oncotarget, 7, 2016
|
|
5YXB
 
 | | A ligand binding to FXR | | Descriptor: | 2-methoxyethyl (2E)-3-phenylprop-2-en-1-yl 2,6-dimethyl-4-(3-nitrophenyl)pyridine-3,5-dicarboxylate, Bile acid receptor, Peptide from Nuclear receptor coactivator 2 | | Authors: | Yi, L, Yong, L. | | Deposit date: | 2017-12-04 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A ligand binding to FXR
To Be Published
|
|
5ZCH
 
 | | Crystal structure of OsPP2C50 I267W:OsPYL/RCAR3 with (+)-ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, MAGNESIUM ION, ... | | Authors: | Lee, S, Han, S. | | Deposit date: | 2018-02-17 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.474 Å) | | Cite: | Comprehensive survey of the VxG Phi L motif of PP2Cs from Oryza sativa reveals the critical role of the fourth position in regulation of ABA responsiveness.
Plant Mol.Biol., 101, 2019
|
|
8D1Y
 
 | |
3FMR
 
 | | Crystal structure of an Encephalitozoon cuniculi methionine aminopeptidase type 2 with angiogenesis inhibitor TNP470 bound | | Descriptor: | (1R,2S,3S,4R)-4-hydroxy-2-methoxy-4-methyl-3-[(2R,3R)-2-methyl-3-(3-methylbut-2-en-1-yl)oxiran-2-yl]cyclohexyl (chloroacetyl)carbamate, FE (III) ION, Methionine aminopeptidase 2, ... | | Authors: | Alvarado, J.J, Russell, M, Zhang, A, Adams, J, Toro, R, Burley, S.K, Weiss, L.M, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-22 | | Release date: | 2009-01-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of a microsporidian methionine aminopeptidase type 2 complexed with fumagillin and TNP-470.
Mol.Biochem.Parasitol., 168, 2009
|
|
6NT4
 
 | | Cryo-EM structure of a human-cockroach hybrid Nav channel bound to alpha-scorpion toxin AaH2. | | Descriptor: | (7E,21R,24S)-27-amino-24-hydroxy-18,24-dioxo-19,23,25-trioxa-24lambda~5~-phosphaheptacos-7-en-21-yl (9Z,12E)-octadeca-9,12-dienoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Clairfeuille, T, Rohou, A, Payandeh, J. | | Deposit date: | 2019-01-28 | | Release date: | 2019-02-20 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of alpha-scorpion toxin action on Na v channels.
Science, 363, 2019
|
|
3FO9
 
 | |
3FP0
 
 | | Structural and Functional Characterization of TRI3 Trichothecene 15-O-acetyltransferase from Fusarium sporotrichioides | | Descriptor: | (3alpha)-15-hydroxy-12,13-epoxytrichothec-9-en-3-yl acetate, 15-O-acetyltransferase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Garvey, G.S, Rayment, I, McCormick, S.P, Alexander, N.J. | | Deposit date: | 2009-01-02 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional characterization of TRI3 trichothecene 15-O-acetyltransferase from Fusarium sporotrichioides
Protein Sci., 18, 2009
|
|
6AA4
 
 | | Crystal structure of MTH1 in complex with alpha-mangostin (cocktail No. 9) | | Descriptor: | 1,3,6-trihydroxy-7-methoxy-2,8-bis(3-methylbut-2-en-1-yl)-9H-xanthen-9-one, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION, ... | | Authors: | Yokoyama, T, Kitakami, R, Mizuguchi, M. | | Deposit date: | 2018-07-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a new class of MTH1 inhibitor by X-ray crystallographic screening.
Eur J Med Chem, 167, 2019
|
|
8E43
 
 | | E. coli 50S ribosome bound to compound streptogramin A analog 3336 | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,16R,23S,26aR)-16-fluoro-14-hydroxy-12-methyl-1,7,22-trioxo-4-(prop-2-en-1-yl)-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl isoquinolin-3-ylcarbamate, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Seiple, I.B, Fraser, J.S. | | Deposit date: | 2022-08-17 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.09 Å) | | Cite: | Streptogramin A analogs
To Be Published
|
|
5ZWH
 
 | | Covalent bond formation between histidine of Vitamin D receptor (VDR) and a full agonist having an ene-ynone group via conjugate addition reaction | | Descriptor: | (2S)-2-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]oct-4,6-diene-3-one, (E,2S)-2-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]oct-6-en-4-yn-3-one, 13-meric peptide from DRIP205 NR2 BOX peptide, ... | | Authors: | Yoshizawa, M, Itoh, T, Anami, Y, Kato, A, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-18 | | Last modified: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands
J. Med. Chem., 61, 2018
|
|
6OOB
 
 | | Human CYP3A4 bound to a suicide substrate | | Descriptor: | 4-{[(2Z,6S)-6,7-dihydroxy-3,7-dimethyloct-2-en-1-yl]oxy}-7H-furo[3,2-g][1]benzopyran-7-one, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2019-04-22 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structural Insights into the Interaction of Cytochrome P450 3A4 with Suicide Substrates: Mibefradil, Azamulin and 6',7'-Dihydroxybergamottin.
Int J Mol Sci, 20, 2019
|
|
5ZWE
 
 | | Covalent bond formation between histidine of Vitamin D receptor (VDR) and a full agonist having a vinyl ketone group via conjugate addition reaction | | Descriptor: | (6R)-6-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]hept-1-en-3-one, 13-meric peptide from DRIP205 NR2 BOX peptide, Vitamin D3 receptor | | Authors: | Yoshizawa, M, Itoh, T, Anami, Y, Kato, A, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-18 | | Last modified: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands
J. Med. Chem., 61, 2018
|
|
8ED7
 
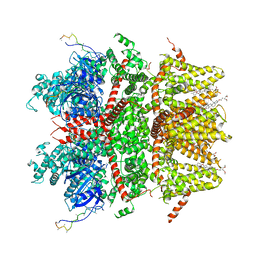 | | cryo-EM structure of TRPM3 ion channel in apo state | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, Transient receptor potential cation channel, subfamily M, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-09-03 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8ED8
 
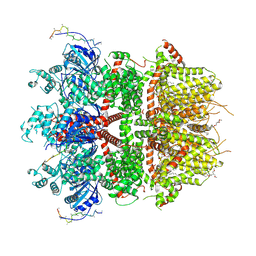 | | cryo-EM structure of TRPM3 ion channel in the presence of PIP2 and PregS, state 1 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-09-03 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
6OBA
 
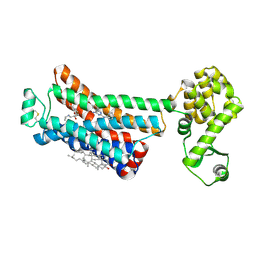 | | The beta2 adrenergic receptor bound to a negative allosteric modulator | | Descriptor: | (2S)-1-[(1-methylethyl)amino]-3-(2-prop-2-en-1-ylphenoxy)propan-2-ol, 6-bromo-N~2~-phenylquinazoline-2,4-diamine, Beta-2 adrenergic receptor,Lysozyme,Beta-2 adrenergic receptor, ... | | Authors: | Liu, X, Stobel, A, Kaindl, J, Dengler, D, ClarK, M, Mahoney, J, Korczynska, M, Matt, R.A, Hubner, H, Xu, X, Stanek, M, Hirata, K, Shoichet, B, Sunahara, R, Gmeiner, R, Kobilka, B.K. | | Deposit date: | 2019-03-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An allosteric modulator binds to a conformational hub in the beta2adrenergic receptor.
Nat.Chem.Biol., 16, 2020
|
|
8ED9
 
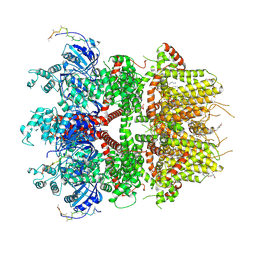 | | cryo-EM structure of TRPM3 ion channel in the presence with PIP2 and PregS, state 2 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-09-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
